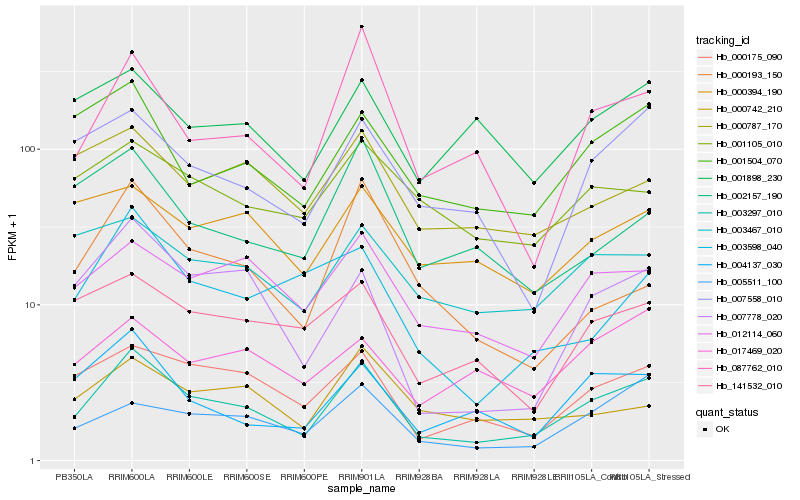
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003297_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |
| 2 |
Hb_000175_090 |
0.1478926561 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_087762_010 |
0.1547771332 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 4 |
Hb_012114_060 |
0.15731219 |
- |
- |
PREDICTED: uncharacterized protein LOC105645814 [Jatropha curcas] |
| 5 |
Hb_007778_020 |
0.1602632036 |
- |
- |
- |
| 6 |
Hb_007558_010 |
0.1615955022 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM9-2 [Jatropha curcas] |
| 7 |
Hb_004137_030 |
0.1619251413 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001504_070 |
0.1620171494 |
- |
- |
PREDICTED: 60S ribosomal protein L7-1 [Jatropha curcas] |
| 9 |
Hb_005511_100 |
0.1671285675 |
- |
- |
PREDICTED: uncharacterized protein LOC105645493 isoform X1 [Jatropha curcas] |
| 10 |
Hb_141532_010 |
0.1743303733 |
- |
- |
PREDICTED: protein SMG9-like [Jatropha curcas] |
| 11 |
Hb_003467_010 |
0.1759125753 |
- |
- |
PREDICTED: probable thiol methyltransferase 2 [Jatropha curcas] |
| 12 |
Hb_000742_210 |
0.1805963816 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 13 |
Hb_003598_040 |
0.1816932346 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-8 [Jatropha curcas] |
| 14 |
Hb_001105_010 |
0.1826457289 |
desease resistance |
Gene Name: AIG1 |
PREDICTED: translocase of chloroplast 33, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_000787_170 |
0.1830098535 |
- |
- |
hypothetical protein POPTR_0005s06400g [Populus trichocarpa] |
| 16 |
Hb_001898_230 |
0.1831811366 |
- |
- |
PREDICTED: KH domain-containing protein At1g09660/At1g09670 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002157_190 |
0.1841020833 |
- |
- |
PREDICTED: mRNA turnover protein 4 homolog [Jatropha curcas] |
| 18 |
Hb_017469_020 |
0.1841811962 |
- |
- |
- |
| 19 |
Hb_000193_150 |
0.1841981673 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100 [Jatropha curcas] |
| 20 |
Hb_000394_190 |
0.1847452658 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |