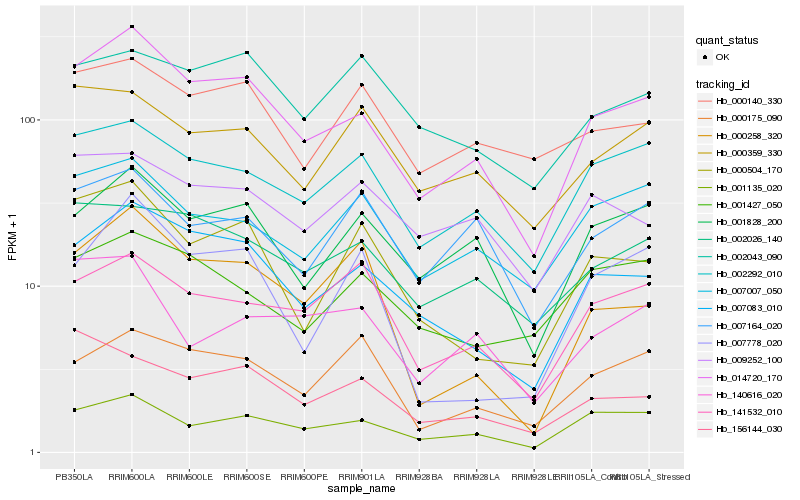
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014720_170 |
0.0 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 2 |
Hb_007083_010 |
0.0879052469 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 13-like, partial [Nelumbo nucifera] |
| 3 |
Hb_001135_020 |
0.1194027049 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 4 |
Hb_001828_200 |
0.1229303826 |
- |
- |
ribosomal protein L13 [Populus trichocarpa] |
| 5 |
Hb_002292_010 |
0.1231118367 |
- |
- |
hypothetical protein CICLE_v10029628mg [Citrus clementina] |
| 6 |
Hb_002026_140 |
0.127359898 |
- |
- |
hypothetical protein POPTR_0006s13340g [Populus trichocarpa] |
| 7 |
Hb_000504_170 |
0.1309159285 |
- |
- |
hypothetical protein B456_006G033900 [Gossypium raimondii] |
| 8 |
Hb_007007_050 |
0.1420375603 |
- |
- |
PREDICTED: ubiquitin-related modifier 1 homolog 2 [Malus domestica] |
| 9 |
Hb_000140_330 |
0.1457287464 |
- |
- |
PREDICTED: uncharacterized protein LOC105642909 [Jatropha curcas] |
| 10 |
Hb_000359_330 |
0.1479968673 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_141532_010 |
0.1495837616 |
- |
- |
PREDICTED: protein SMG9-like [Jatropha curcas] |
| 12 |
Hb_009252_100 |
0.150370193 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 13 |
Hb_000175_090 |
0.150379912 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000258_320 |
0.1512439227 |
- |
- |
- |
| 15 |
Hb_001427_050 |
0.152473367 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 16 |
Hb_156144_030 |
0.1536517289 |
- |
- |
hypothetical protein RCOM_0187400 [Ricinus communis] |
| 17 |
Hb_007164_020 |
0.1552861186 |
- |
- |
hypothetical protein glysoja_025872 [Glycine soja] |
| 18 |
Hb_007778_020 |
0.156340222 |
- |
- |
- |
| 19 |
Hb_002043_090 |
0.1564768421 |
- |
- |
hypothetical protein JCGZ_24811 [Jatropha curcas] |
| 20 |
Hb_140616_020 |
0.1573263044 |
- |
- |
- |