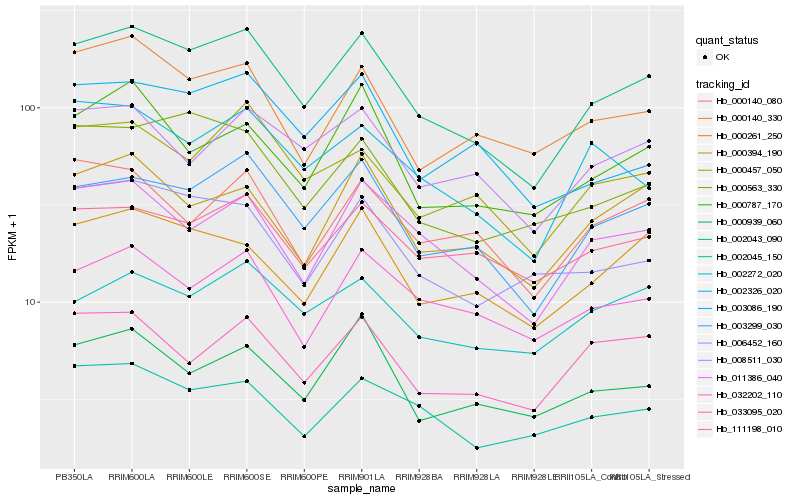
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002043_090 |
0.0 |
- |
- |
hypothetical protein JCGZ_24811 [Jatropha curcas] |
| 2 |
Hb_003299_030 |
0.0583814885 |
- |
- |
PREDICTED: uncharacterized protein At1g27050 isoform X1 [Jatropha curcas] |
| 3 |
Hb_011386_040 |
0.0812252643 |
- |
- |
PREDICTED: uncharacterized protein LOC105639243 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000457_050 |
0.0882303608 |
- |
- |
PREDICTED: tether containing UBX domain for GLUT4 [Jatropha curcas] |
| 5 |
Hb_000261_250 |
0.0900997519 |
- |
- |
elongation factor 1-alpha [Lepidium sativum] |
| 6 |
Hb_000563_330 |
0.0925340675 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_002045_150 |
0.0928515699 |
- |
- |
histone acetyltransferase gcn5, putative [Ricinus communis] |
| 8 |
Hb_000394_190 |
0.0932514176 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_002326_020 |
0.093437986 |
- |
- |
hypothetical protein POPTR_0002s13160g [Populus trichocarpa] |
| 10 |
Hb_008511_030 |
0.094877334 |
- |
- |
guanine nucleotide-binding protein beta, putative [Ricinus communis] |
| 11 |
Hb_000140_330 |
0.0965110233 |
- |
- |
PREDICTED: uncharacterized protein LOC105642909 [Jatropha curcas] |
| 12 |
Hb_003086_190 |
0.0965372814 |
- |
- |
PREDICTED: uncharacterized protein LOC105635794 [Jatropha curcas] |
| 13 |
Hb_000787_170 |
0.096848542 |
- |
- |
hypothetical protein POPTR_0005s06400g [Populus trichocarpa] |
| 14 |
Hb_006452_160 |
0.0968764196 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 15 |
Hb_111198_010 |
0.0972368971 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_033095_020 |
0.0975497695 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002272_020 |
0.0985850685 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |
| 18 |
Hb_032202_110 |
0.0987876929 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 19 |
Hb_000939_060 |
0.0991468783 |
- |
- |
PREDICTED: DNA repair protein REV1 [Jatropha curcas] |
| 20 |
Hb_000140_080 |
0.1020153954 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |