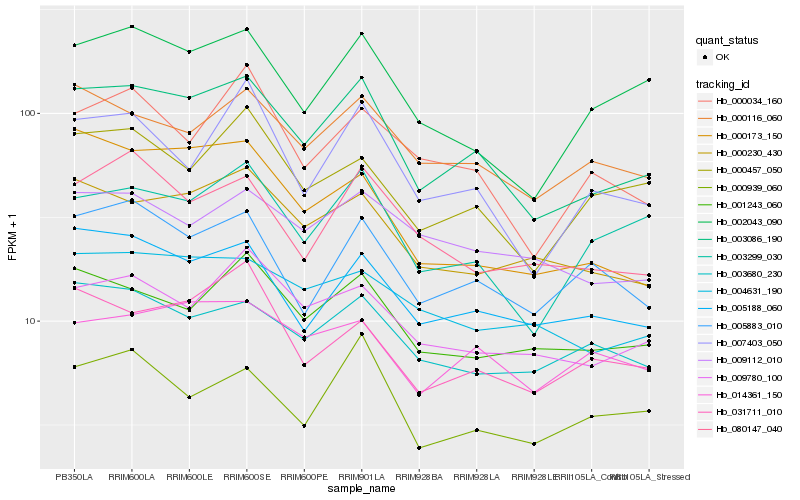
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003086_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635794 [Jatropha curcas] |
| 2 |
Hb_005188_060 |
0.089002346 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 3 |
Hb_000116_060 |
0.0898103744 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 4 |
Hb_000230_430 |
0.090881578 |
- |
- |
catalytic, putative [Ricinus communis] |
| 5 |
Hb_001243_060 |
0.0918422254 |
- |
- |
PREDICTED: probable magnesium transporter NIPA6 [Jatropha curcas] |
| 6 |
Hb_003299_030 |
0.0953328995 |
- |
- |
PREDICTED: uncharacterized protein At1g27050 isoform X1 [Jatropha curcas] |
| 7 |
Hb_009780_100 |
0.0956925298 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 8 |
Hb_002043_090 |
0.0965372814 |
- |
- |
hypothetical protein JCGZ_24811 [Jatropha curcas] |
| 9 |
Hb_007403_050 |
0.0983971324 |
- |
- |
PREDICTED: protein YIPF5 homolog [Jatropha curcas] |
| 10 |
Hb_005883_010 |
0.0988060176 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003680_230 |
0.1003531348 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 12 |
Hb_004631_190 |
0.1005391701 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 13 |
Hb_080147_040 |
0.1011708832 |
- |
- |
hypothetical protein JCGZ_07266 [Jatropha curcas] |
| 14 |
Hb_014361_150 |
0.1016067301 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 15 |
Hb_000457_050 |
0.1025633787 |
- |
- |
PREDICTED: tether containing UBX domain for GLUT4 [Jatropha curcas] |
| 16 |
Hb_000939_060 |
0.1045083571 |
- |
- |
PREDICTED: DNA repair protein REV1 [Jatropha curcas] |
| 17 |
Hb_031711_010 |
0.1049725709 |
- |
- |
PREDICTED: CDPK-related kinase 4-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000173_150 |
0.105129682 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |
| 19 |
Hb_009112_010 |
0.1061527263 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 20 |
Hb_000034_160 |
0.1064673478 |
- |
- |
hypothetical protein EUGRSUZ_F02151 [Eucalyptus grandis] |