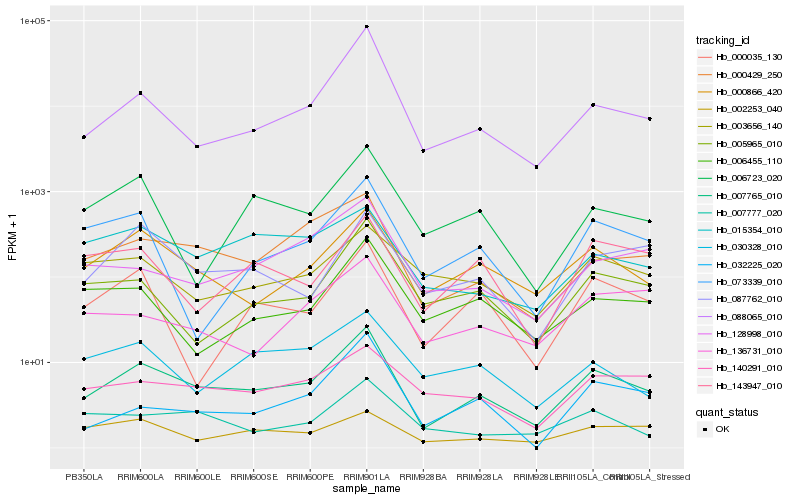
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007765_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000866_420 |
0.1443512086 |
- |
- |
Remorin, putative [Ricinus communis] |
| 3 |
Hb_000429_250 |
0.1586277333 |
- |
- |
hypothetical protein L484_000744 [Morus notabilis] |
| 4 |
Hb_087762_010 |
0.1748651753 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 5 |
Hb_088065_010 |
0.1757220373 |
- |
- |
hypothetical protein glysoja_007934 [Glycine soja] |
| 6 |
Hb_000035_130 |
0.1777673245 |
- |
- |
hypothetical protein Csa_4G652025 [Cucumis sativus] |
| 7 |
Hb_005965_010 |
0.1807645337 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb family transcription factor APL-like [Jatropha curcas] |
| 8 |
Hb_073339_010 |
0.1854614993 |
- |
- |
hypothetical protein 30 [Hevea brasiliensis] |
| 9 |
Hb_006723_020 |
0.1858416129 |
- |
- |
ARF2-B: ADP-ribosylation factor 2-B [Gossypium arboreum] |
| 10 |
Hb_128998_010 |
0.187723634 |
- |
- |
PREDICTED: uncharacterized protein LOC105650916 [Jatropha curcas] |
| 11 |
Hb_140291_010 |
0.1887800413 |
- |
- |
hypothetical protein JCGZ_17222 [Jatropha curcas] |
| 12 |
Hb_007777_020 |
0.1891029332 |
- |
- |
hypothetical protein JCGZ_14410 [Jatropha curcas] |
| 13 |
Hb_006455_110 |
0.1902113436 |
- |
- |
bromodomain-containing protein, putative [Ricinus communis] |
| 14 |
Hb_030328_010 |
0.1921803651 |
- |
- |
hypothetical protein CICLE_v10015936mg [Citrus clementina] |
| 15 |
Hb_015354_010 |
0.1956086277 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 16 |
Hb_143947_010 |
0.1963414354 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X2 [Pyrus x bretschneideri] |
| 17 |
Hb_136731_010 |
0.1995571609 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X4 [Jatropha curcas] |
| 18 |
Hb_032225_020 |
0.2006978489 |
- |
- |
hypothetical protein POPTR_0006s08140g [Populus trichocarpa] |
| 19 |
Hb_002253_040 |
0.2015278591 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g13690 [Jatropha curcas] |
| 20 |
Hb_003656_140 |
0.2021900086 |
- |
- |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Jatropha curcas] |