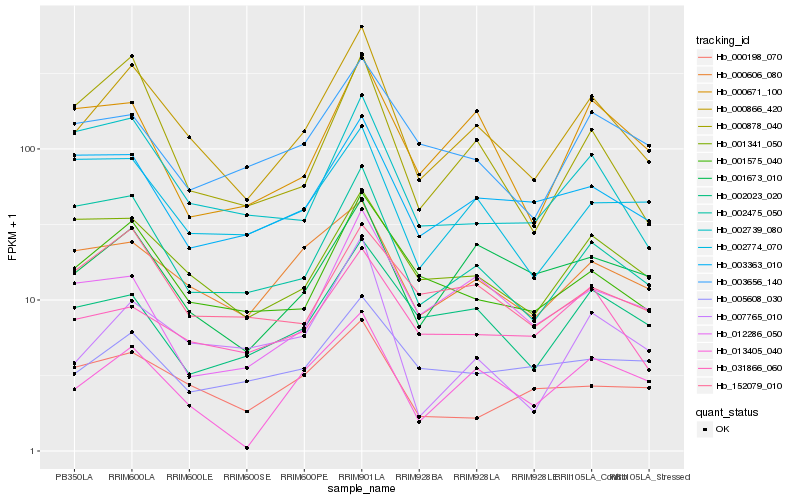
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000866_420 |
0.0 |
- |
- |
Remorin, putative [Ricinus communis] |
| 2 |
Hb_001575_040 |
0.1290901068 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 25 isoform X2 [Jatropha curcas] |
| 3 |
Hb_013405_040 |
0.1365254172 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002475_050 |
0.1435928723 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 5 |
Hb_007765_010 |
0.1443512086 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000606_080 |
0.1444062439 |
- |
- |
PREDICTED: RNA-binding protein Nova-2-like [Malus domestica] |
| 7 |
Hb_001673_010 |
0.146044764 |
- |
- |
PREDICTED: uncharacterized protein LOC105635783 isoform X3 [Jatropha curcas] |
| 8 |
Hb_031866_060 |
0.1520868343 |
- |
- |
PREDICTED: uncharacterized protein LOC105642381 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001341_050 |
0.1554164067 |
- |
- |
ATP synthase D chain, mitochondrial [Theobroma cacao] |
| 10 |
Hb_002023_020 |
0.1593220281 |
- |
- |
PREDICTED: aladin isoform X2 [Jatropha curcas] |
| 11 |
Hb_000878_040 |
0.161087914 |
- |
- |
PREDICTED: uncharacterized protein LOC105635780 isoform X1 [Jatropha curcas] |
| 12 |
Hb_152079_010 |
0.1618498403 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105645015 [Jatropha curcas] |
| 13 |
Hb_002739_080 |
0.1631112525 |
- |
- |
cystathionine beta-lyase, putative [Ricinus communis] |
| 14 |
Hb_003656_140 |
0.163728967 |
- |
- |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003363_010 |
0.1650950417 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000671_100 |
0.1650972869 |
- |
- |
hypothetical protein POPTR_0017s06410g [Populus trichocarpa] |
| 17 |
Hb_005608_030 |
0.1675134739 |
- |
- |
PREDICTED: uncharacterized protein LOC104110884 [Nicotiana tomentosiformis] |
| 18 |
Hb_002774_070 |
0.1683548281 |
- |
- |
hypothetical protein EUGRSUZ_F00307 [Eucalyptus grandis] |
| 19 |
Hb_000198_070 |
0.1692223477 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 7-like [Jatropha curcas] |
| 20 |
Hb_012286_050 |
0.1704712787 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |