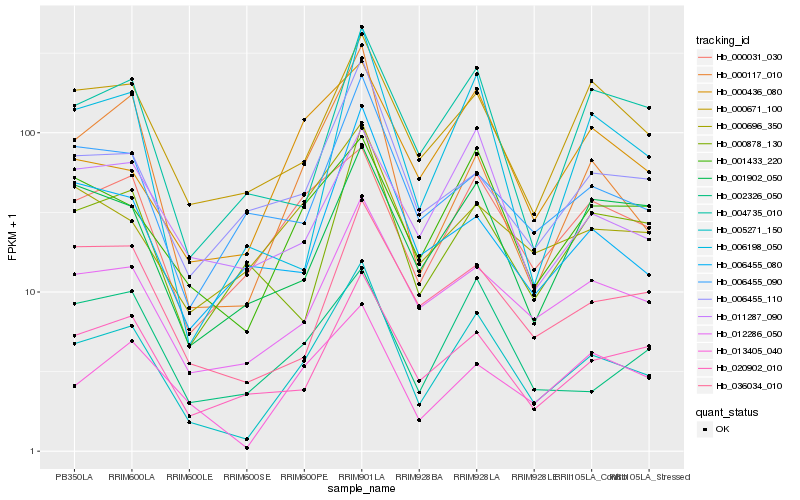
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005271_150 |
0.0 |
- |
- |
PREDICTED: EKC/KEOPS complex subunit Tprkb-like [Malus domestica] |
| 2 |
Hb_012286_050 |
0.1386707856 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 3 |
Hb_000117_010 |
0.1479729642 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like [Malus domestica] |
| 4 |
Hb_020902_010 |
0.1552492694 |
- |
- |
PREDICTED: uncharacterized protein LOC104595551 [Nelumbo nucifera] |
| 5 |
Hb_001433_220 |
0.1579333339 |
- |
- |
protein phosphatase, putative [Ricinus communis] |
| 6 |
Hb_013405_040 |
0.1603674357 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000671_100 |
0.1653921414 |
- |
- |
hypothetical protein POPTR_0017s06410g [Populus trichocarpa] |
| 8 |
Hb_006198_050 |
0.1656580589 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_000031_030 |
0.1691670087 |
- |
- |
PREDICTED: plasma membrane ATPase 4 [Jatropha curcas] |
| 10 |
Hb_006455_110 |
0.1692773277 |
- |
- |
bromodomain-containing protein, putative [Ricinus communis] |
| 11 |
Hb_036034_010 |
0.1694281196 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000696_350 |
0.173115529 |
- |
- |
ARM repeat superfamily protein isoform 2 [Theobroma cacao] |
| 13 |
Hb_006455_090 |
0.1731612541 |
- |
- |
PREDICTED: transcription factor GTE1-like [Jatropha curcas] |
| 14 |
Hb_004735_010 |
0.1736372696 |
- |
- |
PREDICTED: protein arginine N-methyltransferase PRMT10 [Vitis vinifera] |
| 15 |
Hb_000436_080 |
0.1737883517 |
- |
- |
unnamed protein product [Coffea canephora] |
| 16 |
Hb_002326_050 |
0.1775328453 |
- |
- |
PREDICTED: uncharacterized protein LOC105633701 [Jatropha curcas] |
| 17 |
Hb_006455_080 |
0.1788127274 |
- |
- |
PREDICTED: transcription factor GTE6-like isoform X2 [Nelumbo nucifera] |
| 18 |
Hb_000878_130 |
0.179271662 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_011287_090 |
0.1802727588 |
- |
- |
PREDICTED: GDP-mannose transporter GONST3-like [Populus euphratica] |
| 20 |
Hb_001902_050 |
0.180625689 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105648450 [Jatropha curcas] |