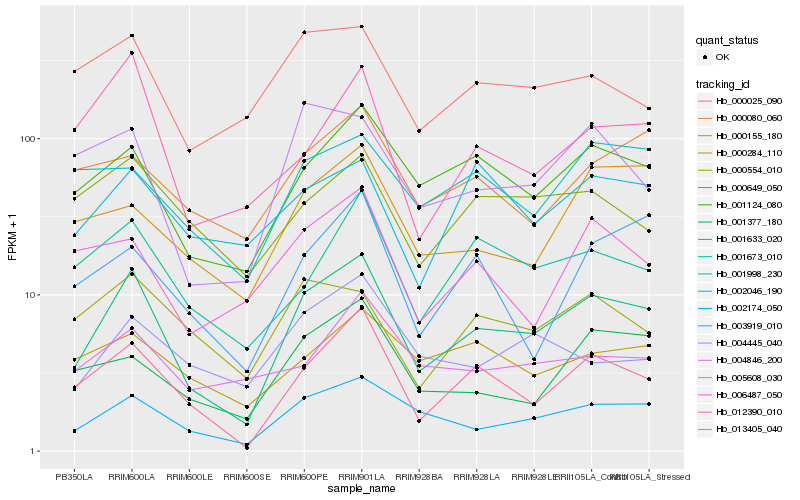
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001633_020 |
0.0 |
- |
- |
F22C12.10, putative isoform 2 [Theobroma cacao] |
| 2 |
Hb_013405_040 |
0.1297395687 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002174_050 |
0.1365904228 |
- |
- |
- |
| 4 |
Hb_001124_080 |
0.145413309 |
- |
- |
RecName: Full=Acyl carrier protein 2, chloroplastic; Short=ACP; Flags: Precursor [Cuphea lanceolata] |
| 5 |
Hb_001673_010 |
0.1731252833 |
- |
- |
PREDICTED: uncharacterized protein LOC105635783 isoform X3 [Jatropha curcas] |
| 6 |
Hb_001377_180 |
0.176872124 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1 [Jatropha curcas] |
| 7 |
Hb_004846_200 |
0.1802202389 |
- |
- |
PREDICTED: shikimate O-hydroxycinnamoyltransferase [Jatropha curcas] |
| 8 |
Hb_012390_010 |
0.1825268079 |
- |
- |
pectate lyase 1 [Hevea brasiliensis] |
| 9 |
Hb_003919_010 |
0.1887215935 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, mitochondrial isoform X1 [Jatropha curcas] |
| 10 |
Hb_000155_180 |
0.189718937 |
- |
- |
PREDICTED: 28 kDa heat- and acid-stable phosphoprotein [Jatropha curcas] |
| 11 |
Hb_006487_050 |
0.1897499478 |
- |
- |
hypothetical protein PHAVU_005G049600g [Phaseolus vulgaris] |
| 12 |
Hb_000025_090 |
0.1905818661 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 13 |
Hb_000080_060 |
0.1923204649 |
- |
- |
RNA polymerase I, II and III subunit RPB8, partial [Manihot esculenta] |
| 14 |
Hb_004445_040 |
0.1941935397 |
- |
- |
PREDICTED: uncharacterized protein LOC105639134 isoform X5 [Jatropha curcas] |
| 15 |
Hb_001998_230 |
0.1944657648 |
- |
- |
PREDICTED: coatomer subunit epsilon-1 [Jatropha curcas] |
| 16 |
Hb_002046_190 |
0.1947684372 |
- |
- |
calcium-binding family protein [Populus trichocarpa] |
| 17 |
Hb_000284_110 |
0.1954396923 |
- |
- |
PREDICTED: bromodomain-containing protein 3 [Jatropha curcas] |
| 18 |
Hb_005608_030 |
0.1957717131 |
- |
- |
PREDICTED: uncharacterized protein LOC104110884 [Nicotiana tomentosiformis] |
| 19 |
Hb_000554_010 |
0.1958597345 |
- |
- |
transporter, putative [Ricinus communis] |
| 20 |
Hb_000649_050 |
0.195867533 |
- |
- |
PREDICTED: microtubule-associated protein 70-2-like isoform X1 [Jatropha curcas] |