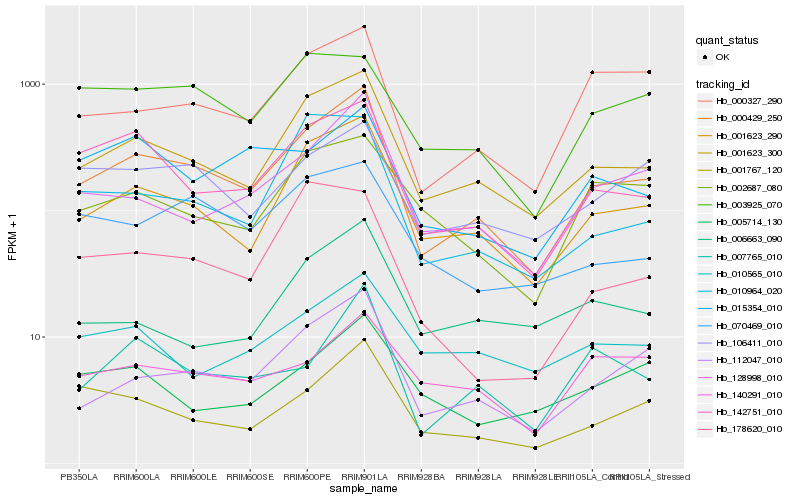
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000429_250 |
0.0 |
- |
- |
hypothetical protein L484_000744 [Morus notabilis] |
| 2 |
Hb_001623_300 |
0.0891733521 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13-B [Jatropha curcas] |
| 3 |
Hb_001623_290 |
0.0955048864 |
- |
- |
unknown [Populus trichocarpa] |
| 4 |
Hb_112047_010 |
0.1214445439 |
- |
- |
- |
| 5 |
Hb_142751_010 |
0.1323648115 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_128998_010 |
0.1352640795 |
- |
- |
PREDICTED: uncharacterized protein LOC105650916 [Jatropha curcas] |
| 7 |
Hb_001767_120 |
0.1466624179 |
- |
- |
- |
| 8 |
Hb_000327_290 |
0.1522443375 |
- |
- |
Histone H4 [Triticum urartu] |
| 9 |
Hb_007765_010 |
0.1586277333 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_106411_010 |
0.1597674282 |
- |
- |
Ribosomal protein S25 family protein [Theobroma cacao] |
| 11 |
Hb_010964_020 |
0.1653468607 |
- |
- |
RNA helicase [Arabidopsis thaliana] |
| 12 |
Hb_070469_010 |
0.1662112879 |
- |
- |
hypothetical protein POPTR_0002s21850g [Populus trichocarpa] |
| 13 |
Hb_002687_080 |
0.1710843682 |
- |
- |
hypothetical protein JCGZ_06535 [Jatropha curcas] |
| 14 |
Hb_015354_010 |
0.1731492888 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 15 |
Hb_003925_070 |
0.1777941789 |
- |
- |
ATOZI1, putative [Ricinus communis] |
| 16 |
Hb_010565_010 |
0.179469347 |
- |
- |
PREDICTED: N-acetylglucosaminyl-phosphatidylinositol de-N-acetylase-like isoform X2 [Populus euphratica] |
| 17 |
Hb_006663_090 |
0.1809038684 |
- |
- |
hypothetical protein B456_010G1255001, partial [Gossypium raimondii] |
| 18 |
Hb_178620_010 |
0.181551342 |
- |
- |
ADP-ribosylation factor, arf, putative [Ricinus communis] |
| 19 |
Hb_140291_010 |
0.18168019 |
- |
- |
hypothetical protein JCGZ_17222 [Jatropha curcas] |
| 20 |
Hb_005714_130 |
0.1828439738 |
transcription factor |
TF Family: C2H2 |
unknown [Populus trichocarpa x Populus deltoides] |