| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
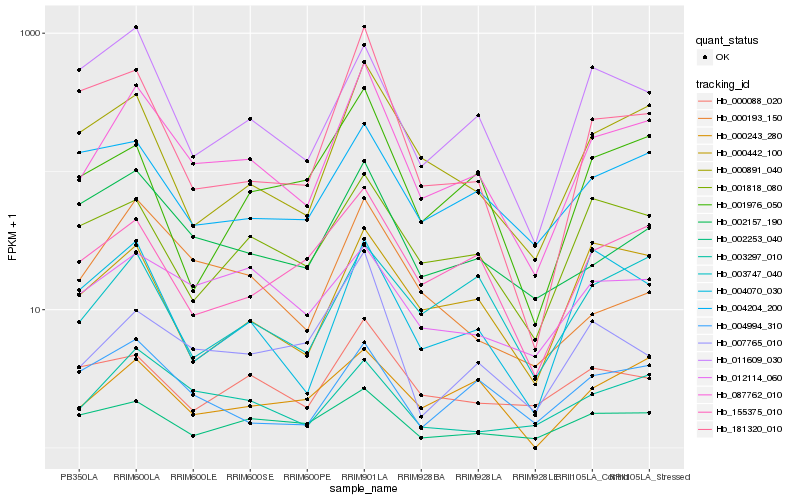
Hb_087762_010 |
0.0 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 2 |
Hb_000891_040 |
0.1499727714 |
- |
- |
hypothetical protein JCGZ_11252 [Jatropha curcas] |
| 3 |
Hb_003297_010 |
0.1547771332 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |
| 4 |
Hb_155375_010 |
0.1650332105 |
- |
- |
hypothetical protein JCGZ_23388 [Jatropha curcas] |
| 5 |
Hb_002157_190 |
0.1699935048 |
- |
- |
PREDICTED: mRNA turnover protein 4 homolog [Jatropha curcas] |
| 6 |
Hb_002253_040 |
0.1719917629 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g13690 [Jatropha curcas] |
| 7 |
Hb_007765_010 |
0.1748651753 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001818_080 |
0.1751785619 |
- |
- |
PREDICTED: F-box protein At3g58530 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003747_040 |
0.1777773641 |
- |
- |
- |
| 10 |
Hb_000442_100 |
0.17838688 |
- |
- |
PREDICTED: staphylococcal-like nuclease CAN2 [Jatropha curcas] |
| 11 |
Hb_000193_150 |
0.1798393274 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100 [Jatropha curcas] |
| 12 |
Hb_000088_020 |
0.1812351774 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_10946 [Jatropha curcas] |
| 13 |
Hb_000243_280 |
0.1812818833 |
- |
- |
- |
| 14 |
Hb_181320_010 |
0.1846932003 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Jatropha curcas] |
| 15 |
Hb_004070_030 |
0.1853456499 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase, mitochondrial isoform X2 [Jatropha curcas] |
| 16 |
Hb_001976_050 |
0.1856870845 |
- |
- |
PREDICTED: protein SKIP34 [Jatropha curcas] |
| 17 |
Hb_004994_310 |
0.1870874053 |
- |
- |
Kiwellin, putative [Ricinus communis] |
| 18 |
Hb_004204_200 |
0.1883131714 |
- |
- |
PREDICTED: LYR motif-containing protein 4 [Jatropha curcas] |
| 19 |
Hb_012114_060 |
0.1893262386 |
- |
- |
PREDICTED: uncharacterized protein LOC105645814 [Jatropha curcas] |
| 20 |
Hb_011609_030 |
0.1895515269 |
- |
- |
conserved hypothetical protein 13 [Hevea brasiliensis] |