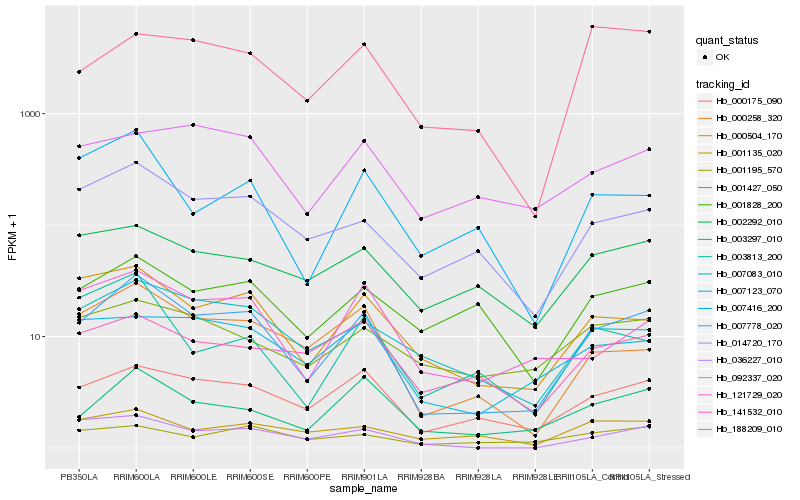
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007778_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000175_090 |
0.1463878607 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000504_170 |
0.149876047 |
- |
- |
hypothetical protein B456_006G033900 [Gossypium raimondii] |
| 4 |
Hb_000258_320 |
0.1545907082 |
- |
- |
- |
| 5 |
Hb_014720_170 |
0.156340222 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 6 |
Hb_007083_010 |
0.1591904756 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 13-like, partial [Nelumbo nucifera] |
| 7 |
Hb_003297_010 |
0.1602632036 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |
| 8 |
Hb_036227_010 |
0.1663399245 |
- |
- |
- |
| 9 |
Hb_121729_020 |
0.1747672508 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_007123_070 |
0.1773877344 |
- |
- |
- |
| 11 |
Hb_007416_200 |
0.1824291065 |
- |
- |
PREDICTED: putative peptidyl-tRNA hydrolase PTRHD1 [Jatropha curcas] |
| 12 |
Hb_141532_010 |
0.1890699898 |
- |
- |
PREDICTED: protein SMG9-like [Jatropha curcas] |
| 13 |
Hb_001828_200 |
0.1904580763 |
- |
- |
ribosomal protein L13 [Populus trichocarpa] |
| 14 |
Hb_188209_010 |
0.1924864026 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 7B, mitochondrial-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_001427_050 |
0.1927179409 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 16 |
Hb_002292_010 |
0.1929273033 |
- |
- |
hypothetical protein CICLE_v10029628mg [Citrus clementina] |
| 17 |
Hb_092337_020 |
0.1929775437 |
- |
- |
hypothetical protein B456_013G264900 [Gossypium raimondii] |
| 18 |
Hb_001135_020 |
0.1939468018 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 19 |
Hb_003813_200 |
0.1956000961 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 20 |
Hb_001195_570 |
0.1967504402 |
- |
- |
PREDICTED: peroxisomal membrane protein 11A [Jatropha curcas] |