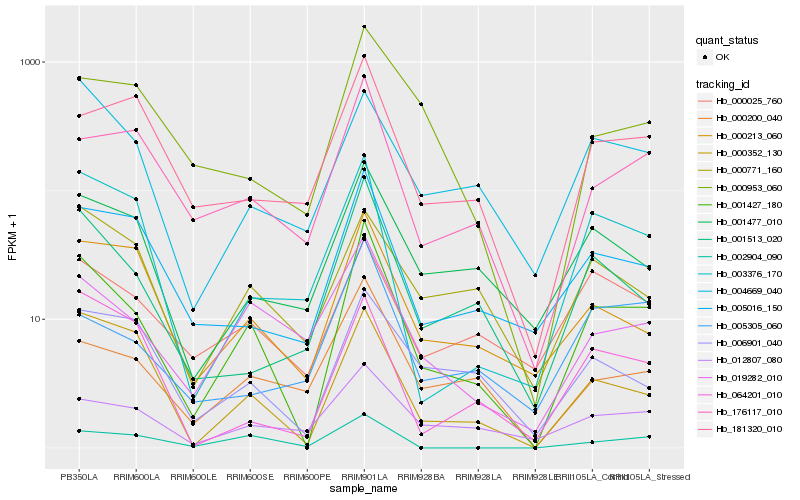
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001427_180 |
0.0 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At4g29420 [Jatropha curcas] |
| 2 |
Hb_001513_020 |
0.1930403753 |
- |
- |
PREDICTED: methionine aminopeptidase 1A isoform X1 [Jatropha curcas] |
| 3 |
Hb_000352_130 |
0.1943042663 |
- |
- |
hypothetical protein CISIN_1g0419111mg, partial [Citrus sinensis] |
| 4 |
Hb_019282_010 |
0.1961856251 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 5 |
Hb_176117_010 |
0.2067411671 |
- |
- |
Cold-inducible RNA-binding protein [Morus notabilis] |
| 6 |
Hb_005016_150 |
0.2085137062 |
- |
- |
- |
| 7 |
Hb_002904_090 |
0.2098429337 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71420-like [Vitis vinifera] |
| 8 |
Hb_000213_060 |
0.2102000998 |
- |
- |
PREDICTED: RRP12-like protein isoform X1 [Jatropha curcas] |
| 9 |
Hb_000200_040 |
0.2111427944 |
- |
- |
- |
| 10 |
Hb_003376_170 |
0.2137144196 |
- |
- |
PREDICTED: uncharacterized protein LOC105642198 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001477_010 |
0.2141345618 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 13-like [Cicer arietinum] |
| 12 |
Hb_012807_080 |
0.2145055986 |
- |
- |
hypothetical protein CICLE_v10028099mg [Citrus clementina] |
| 13 |
Hb_004669_040 |
0.2213909652 |
- |
- |
ATPase inhibitor, putative [Ricinus communis] |
| 14 |
Hb_006901_040 |
0.2224263311 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000025_760 |
0.2252148397 |
- |
- |
- |
| 16 |
Hb_000953_060 |
0.2295332451 |
- |
- |
PREDICTED: putative phytosulfokines 6 [Vitis vinifera] |
| 17 |
Hb_181320_010 |
0.2295997518 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Jatropha curcas] |
| 18 |
Hb_000771_160 |
0.2309402355 |
- |
- |
PREDICTED: abscisic acid receptor PYL8 [Jatropha curcas] |
| 19 |
Hb_064201_010 |
0.2313794912 |
- |
- |
hypothetical protein RCOM_1685970 [Ricinus communis] |
| 20 |
Hb_005305_060 |
0.2321330594 |
- |
- |
hypothetical protein MIMGU_mgv1a015111mg [Erythranthe guttata] |