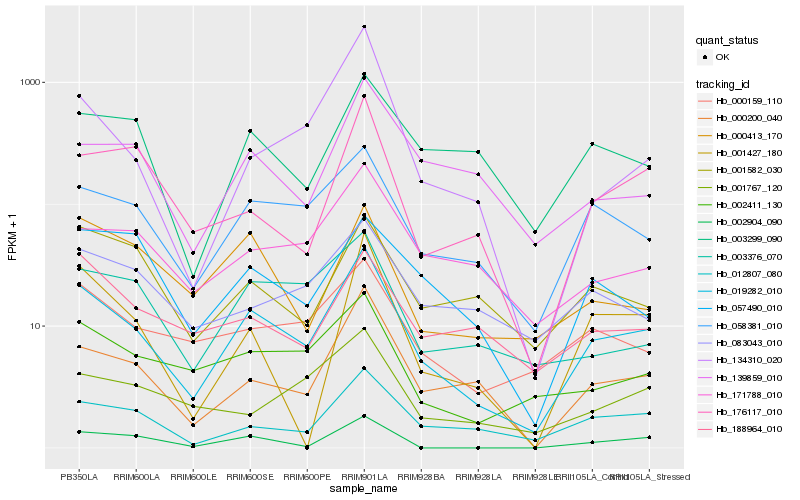
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019282_010 |
0.0 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 2 |
Hb_058381_010 |
0.1541208125 |
- |
- |
hypothetical protein JCGZ_13280 [Jatropha curcas] |
| 3 |
Hb_000200_040 |
0.1606699836 |
- |
- |
- |
| 4 |
Hb_012807_080 |
0.1681699158 |
- |
- |
hypothetical protein CICLE_v10028099mg [Citrus clementina] |
| 5 |
Hb_002411_130 |
0.1798777398 |
- |
- |
PREDICTED: cytosolic sulfotransferase 15-like [Populus euphratica] |
| 6 |
Hb_000159_110 |
0.1812630263 |
- |
- |
PREDICTED: thioredoxin-like protein YLS8 isoform X2 [Malus domestica] |
| 7 |
Hb_003376_070 |
0.1936533844 |
- |
- |
hypothetical protein B456_009G419800 [Gossypium raimondii] |
| 8 |
Hb_171788_010 |
0.1936606362 |
- |
- |
hypothetical protein CISIN_1g007892mg [Citrus sinensis] |
| 9 |
Hb_176117_010 |
0.1960000946 |
- |
- |
Cold-inducible RNA-binding protein [Morus notabilis] |
| 10 |
Hb_001427_180 |
0.1961856251 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At4g29420 [Jatropha curcas] |
| 11 |
Hb_139859_010 |
0.1993434935 |
- |
- |
PREDICTED: cell division control protein 48 homolog D-like [Nicotiana sylvestris] |
| 12 |
Hb_003299_090 |
0.2000858123 |
- |
- |
hypothetical protein PRUPE_ppa011647mg [Prunus persica] |
| 13 |
Hb_000413_170 |
0.2020637149 |
- |
- |
PREDICTED: probable histone H2A.3 [Jatropha curcas] |
| 14 |
Hb_134310_020 |
0.2029441341 |
- |
- |
PREDICTED: putative cytochrome c oxidase subunit 5C-4-like [Citrus sinensis] |
| 15 |
Hb_001582_030 |
0.204706736 |
- |
- |
PREDICTED: uncharacterized protein LOC105639462 [Jatropha curcas] |
| 16 |
Hb_002904_090 |
0.2050988856 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71420-like [Vitis vinifera] |
| 17 |
Hb_057490_010 |
0.20762455 |
- |
- |
PREDICTED: mannose-1-phosphate guanyltransferase alpha [Jatropha curcas] |
| 18 |
Hb_001767_120 |
0.2077343822 |
- |
- |
- |
| 19 |
Hb_188964_010 |
0.2084328694 |
- |
- |
maintenance of killer 16 (mak16) protein, putative [Ricinus communis] |
| 20 |
Hb_083043_010 |
0.210585332 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |