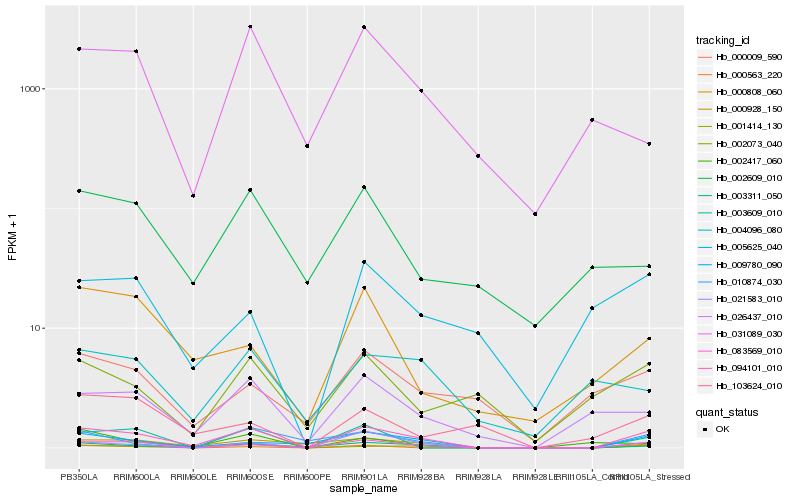
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_094101_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_18414 [Jatropha curcas] |
| 2 |
Hb_083569_010 |
0.160659514 |
- |
- |
- |
| 3 |
Hb_001414_130 |
0.1923579662 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 4 |
Hb_003311_050 |
0.1934102112 |
- |
- |
PREDICTED: two-pore potassium channel 1-like [Jatropha curcas] |
| 5 |
Hb_009780_090 |
0.2279348855 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g66345, mitochondrial [Jatropha curcas] |
| 6 |
Hb_021583_010 |
0.2367027911 |
- |
- |
- |
| 7 |
Hb_000928_150 |
0.244978284 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003609_010 |
0.2571745687 |
- |
- |
- |
| 9 |
Hb_000563_220 |
0.2612528285 |
- |
- |
hypothetical protein VITISV_004111 [Vitis vinifera] |
| 10 |
Hb_026437_010 |
0.2615295557 |
- |
- |
- |
| 11 |
Hb_010874_030 |
0.2627090111 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_10946 [Jatropha curcas] |
| 12 |
Hb_002417_060 |
0.2848172184 |
- |
- |
Cell division cycle protein, putative [Ricinus communis] |
| 13 |
Hb_000808_060 |
0.2922075725 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 16-like [Jatropha curcas] |
| 14 |
Hb_002073_040 |
0.3003669622 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004096_080 |
0.302262118 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 16 |
Hb_031089_030 |
0.3033063254 |
- |
- |
ubiquitin-conjugating enzyme E2 28 [Arabidopsis thaliana] |
| 17 |
Hb_005625_040 |
0.3038059882 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 18 |
Hb_000009_590 |
0.3041125914 |
- |
- |
PREDICTED: prolyl endopeptidase-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_103624_010 |
0.3078431603 |
- |
- |
PREDICTED: protein catecholamines up-like [Jatropha curcas] |
| 20 |
Hb_002609_010 |
0.3079443113 |
- |
- |
PREDICTED: uncharacterized protein LOC105630866 [Jatropha curcas] |