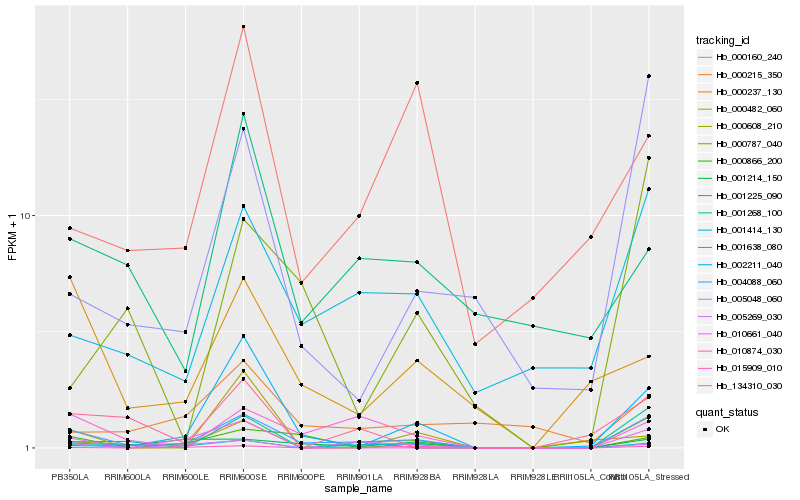
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004088_060 |
0.0 |
- |
- |
hypothetical protein POPTR_0012s01875g [Populus trichocarpa] |
| 2 |
Hb_001414_130 |
0.2660357312 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 3 |
Hb_001638_080 |
0.2689083647 |
- |
- |
PREDICTED: uncharacterized protein LOC105642789 [Jatropha curcas] |
| 4 |
Hb_010874_030 |
0.2800594107 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_10946 [Jatropha curcas] |
| 5 |
Hb_005048_060 |
0.2982016819 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001214_150 |
0.3056584287 |
- |
- |
PREDICTED: uncharacterized protein LOC102624288 [Citrus sinensis] |
| 7 |
Hb_015909_010 |
0.312549103 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_000866_200 |
0.3156406339 |
- |
- |
- |
| 9 |
Hb_000482_060 |
0.3235881384 |
transcription factor |
TF Family: LOB |
hypothetical protein JCGZ_07664 [Jatropha curcas] |
| 10 |
Hb_000237_130 |
0.3260247772 |
- |
- |
- |
| 11 |
Hb_001268_100 |
0.3315942132 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein KNUCKLES [Jatropha curcas] |
| 12 |
Hb_005269_030 |
0.3379309745 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 13 |
Hb_000787_040 |
0.3385798488 |
- |
- |
PREDICTED: uncharacterized protein LOC105115900 isoform X1 [Populus euphratica] |
| 14 |
Hb_002211_040 |
0.3405466019 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.5 [Jatropha curcas] |
| 15 |
Hb_134310_030 |
0.341207911 |
- |
- |
hypothetical protein JCGZ_10790 [Jatropha curcas] |
| 16 |
Hb_001225_090 |
0.3433009029 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_000160_240 |
0.3445793329 |
- |
- |
PREDICTED: uncharacterized protein At1g66480 [Jatropha curcas] |
| 18 |
Hb_010661_040 |
0.3461071372 |
- |
- |
hypothetical protein POPTR_0021s00470g [Populus trichocarpa] |
| 19 |
Hb_000608_210 |
0.3474325838 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein FASCIATED EAR2 [Gossypium raimondii] |
| 20 |
Hb_000215_350 |
0.3476533454 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |