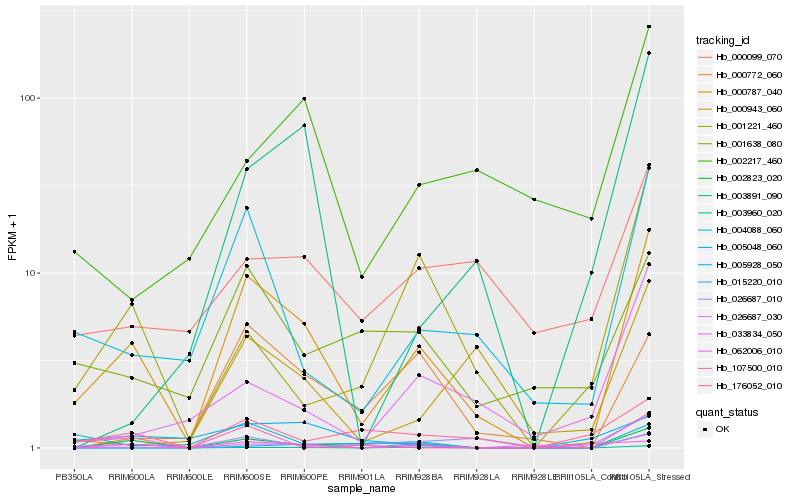
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000787_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105115900 isoform X1 [Populus euphratica] |
| 2 |
Hb_002823_020 |
0.2488973253 |
- |
- |
- |
| 3 |
Hb_005048_060 |
0.2505804267 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001638_080 |
0.2999652997 |
- |
- |
PREDICTED: uncharacterized protein LOC105642789 [Jatropha curcas] |
| 5 |
Hb_000772_060 |
0.3053520535 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_176052_010 |
0.3113139323 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 7 |
Hb_026687_030 |
0.3132392014 |
- |
- |
hypothetical protein AMTR_s00038p00220080 [Amborella trichopoda] |
| 8 |
Hb_026687_010 |
0.3201819761 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Eucalyptus grandis] |
| 9 |
Hb_003960_020 |
0.3250759981 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 10 |
Hb_000099_070 |
0.3279334839 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 11 |
Hb_001221_460 |
0.3282466896 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 12 |
Hb_033834_050 |
0.3341732992 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000943_060 |
0.338336562 |
- |
- |
PREDICTED: uncharacterized protein LOC105635589 [Jatropha curcas] |
| 14 |
Hb_004088_060 |
0.3385798488 |
- |
- |
hypothetical protein POPTR_0012s01875g [Populus trichocarpa] |
| 15 |
Hb_002217_460 |
0.3400407101 |
- |
- |
PREDICTED: transcription factor bHLH147-like [Jatropha curcas] |
| 16 |
Hb_003891_090 |
0.341358524 |
- |
- |
- |
| 17 |
Hb_107500_010 |
0.3479443951 |
- |
- |
- |
| 18 |
Hb_005928_050 |
0.3493853485 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 19 |
Hb_015220_010 |
0.3525929534 |
- |
- |
PREDICTED: ribonuclease 3-like protein 3 [Jatropha curcas] |
| 20 |
Hb_062006_010 |
0.3526365413 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |