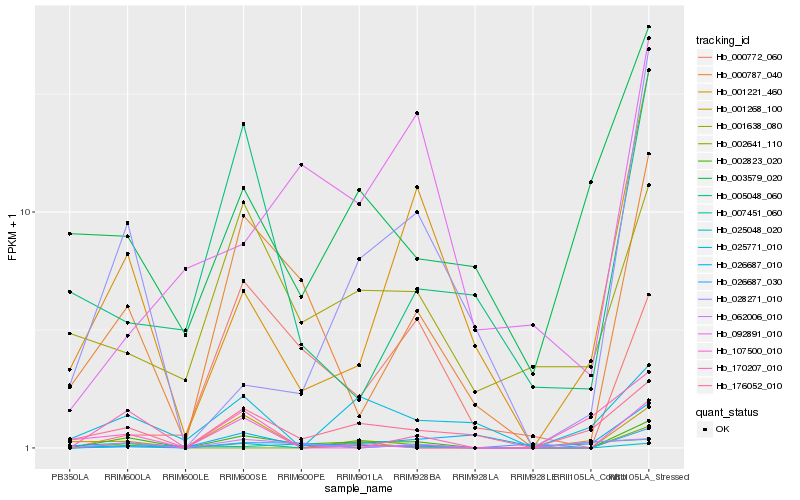
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002823_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000787_040 |
0.2488973253 |
- |
- |
PREDICTED: uncharacterized protein LOC105115900 isoform X1 [Populus euphratica] |
| 3 |
Hb_176052_010 |
0.303806515 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 4 |
Hb_001221_460 |
0.336692916 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 5 |
Hb_001638_080 |
0.341075985 |
- |
- |
PREDICTED: uncharacterized protein LOC105642789 [Jatropha curcas] |
| 6 |
Hb_028271_010 |
0.3490047051 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Populus euphratica] |
| 7 |
Hb_000772_060 |
0.3558644371 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_107500_010 |
0.3582273276 |
- |
- |
- |
| 9 |
Hb_025771_010 |
0.3673206397 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 10 |
Hb_026687_010 |
0.3700901999 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Eucalyptus grandis] |
| 11 |
Hb_001268_100 |
0.3736283055 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein KNUCKLES [Jatropha curcas] |
| 12 |
Hb_092891_010 |
0.3778181342 |
- |
- |
2-nonaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase [Theobroma cacao] |
| 13 |
Hb_026687_030 |
0.3796999624 |
- |
- |
hypothetical protein AMTR_s00038p00220080 [Amborella trichopoda] |
| 14 |
Hb_025048_020 |
0.3809937293 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_005048_060 |
0.3823334004 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_170207_010 |
0.3835878997 |
- |
- |
hypothetical protein JCGZ_01992 [Jatropha curcas] |
| 17 |
Hb_003579_020 |
0.3838015545 |
- |
- |
PREDICTED: probable protein phosphatase 2C 9 [Populus euphratica] |
| 18 |
Hb_007451_060 |
0.3844409364 |
- |
- |
BAHD family acyltransferase, clade V, putative [Theobroma cacao] |
| 19 |
Hb_002641_110 |
0.3857324694 |
- |
- |
PREDICTED: uncharacterized protein LOC103448574 [Malus domestica] |
| 20 |
Hb_062006_010 |
0.3880599511 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |