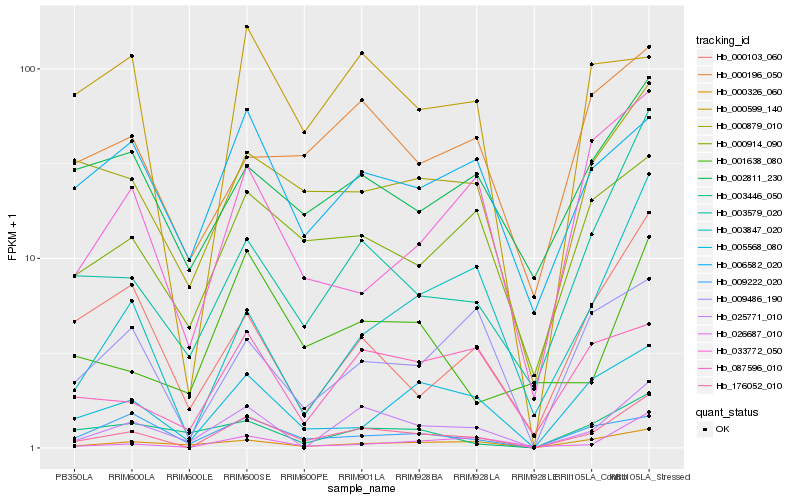
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_176052_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 2 |
Hb_025771_010 |
0.1894535824 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 3 |
Hb_003579_020 |
0.199915555 |
- |
- |
PREDICTED: probable protein phosphatase 2C 9 [Populus euphratica] |
| 4 |
Hb_000326_060 |
0.2074048885 |
- |
- |
PREDICTED: uncharacterized protein LOC104214925 [Nicotiana sylvestris] |
| 5 |
Hb_005568_080 |
0.2113290339 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000879_010 |
0.2160630568 |
- |
- |
PREDICTED: uncharacterized protein LOC105630824 [Jatropha curcas] |
| 7 |
Hb_000914_090 |
0.2256424115 |
- |
- |
PREDICTED: uncharacterized protein LOC105648257 [Jatropha curcas] |
| 8 |
Hb_026687_010 |
0.2324768261 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Eucalyptus grandis] |
| 9 |
Hb_000103_060 |
0.2357706636 |
- |
- |
hypothetical protein JCGZ_02206 [Jatropha curcas] |
| 10 |
Hb_002811_230 |
0.2357841494 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM23-1 [Jatropha curcas] |
| 11 |
Hb_009486_190 |
0.2379030364 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_033772_050 |
0.2379733537 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein A-like [Jatropha curcas] |
| 13 |
Hb_003847_020 |
0.2381140666 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_001638_080 |
0.2411788355 |
- |
- |
PREDICTED: uncharacterized protein LOC105642789 [Jatropha curcas] |
| 15 |
Hb_003446_050 |
0.2412363896 |
- |
- |
hypothetical protein B456_002G034000, partial [Gossypium raimondii] |
| 16 |
Hb_009222_020 |
0.2413678909 |
- |
- |
hypothetical protein AALP_AA5G087000 [Arabis alpina] |
| 17 |
Hb_000196_050 |
0.244463433 |
- |
- |
hypothetical protein JCGZ_20265 [Jatropha curcas] |
| 18 |
Hb_087596_010 |
0.2446326059 |
- |
- |
PREDICTED: disease resistance protein RPS4-like [Jatropha curcas] |
| 19 |
Hb_000599_140 |
0.2450067287 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 4-like [Populus euphratica] |
| 20 |
Hb_006582_020 |
0.245647056 |
- |
- |
Zinc finger protein ZFPM1 [Theobroma cacao] |