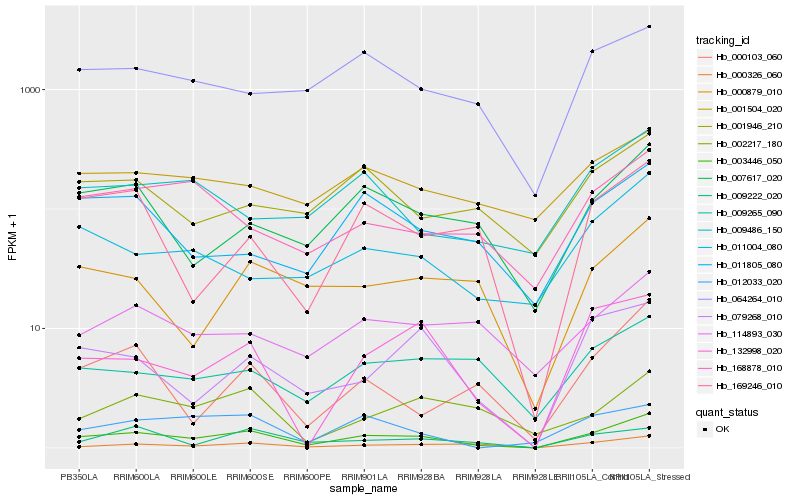
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003446_050 |
0.0 |
- |
- |
hypothetical protein B456_002G034000, partial [Gossypium raimondii] |
| 2 |
Hb_000326_060 |
0.1885146009 |
- |
- |
PREDICTED: uncharacterized protein LOC104214925 [Nicotiana sylvestris] |
| 3 |
Hb_132998_020 |
0.1912147755 |
- |
- |
acyl carrier protein 1, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_007617_020 |
0.195016449 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_079268_010 |
0.1988590503 |
- |
- |
- |
| 6 |
Hb_012033_020 |
0.1999134531 |
- |
- |
- |
| 7 |
Hb_064264_010 |
0.201944561 |
- |
- |
hypothetical protein [Zea mays] |
| 8 |
Hb_011004_080 |
0.206741658 |
- |
- |
PREDICTED: 60S ribosomal protein L37-3-like [Cucumis sativus] |
| 9 |
Hb_000879_010 |
0.2091925967 |
- |
- |
PREDICTED: uncharacterized protein LOC105630824 [Jatropha curcas] |
| 10 |
Hb_009265_090 |
0.2105357975 |
- |
- |
PREDICTED: uncharacterized protein LOC105649618 [Jatropha curcas] |
| 11 |
Hb_168878_010 |
0.2107884657 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 12 |
Hb_169246_010 |
0.2115541654 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_114893_030 |
0.2146024713 |
- |
- |
PREDICTED: ribosome-recycling factor, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000103_060 |
0.2147861972 |
- |
- |
hypothetical protein JCGZ_02206 [Jatropha curcas] |
| 15 |
Hb_009222_020 |
0.2147892633 |
- |
- |
hypothetical protein AALP_AA5G087000 [Arabis alpina] |
| 16 |
Hb_011805_080 |
0.2148350075 |
- |
- |
PREDICTED: uncharacterized protein LOC100242357 [Vitis vinifera] |
| 17 |
Hb_002217_180 |
0.2149298647 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB108-like [Populus euphratica] |
| 18 |
Hb_009486_150 |
0.2153832126 |
- |
- |
60S ribosomal protein L37a-2 [Medicago truncatula] |
| 19 |
Hb_001504_020 |
0.2154179885 |
- |
- |
40S ribosomal protein S12A [Hevea brasiliensis] |
| 20 |
Hb_001946_210 |
0.2167691715 |
- |
- |
ribosomal protein l7ae, putative [Ricinus communis] |