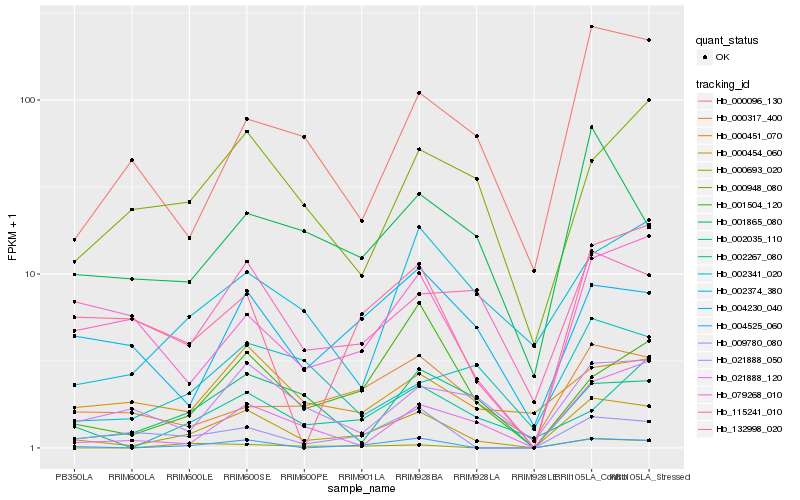
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000454_060 |
0.0 |
- |
- |
Uncharacterized protein TCM_006778 [Theobroma cacao] |
| 2 |
Hb_004525_060 |
0.1942050004 |
- |
- |
- |
| 3 |
Hb_132998_020 |
0.2352627332 |
- |
- |
acyl carrier protein 1, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_021888_050 |
0.2400692544 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 5 |
Hb_001504_120 |
0.2442776509 |
- |
- |
- |
| 6 |
Hb_009780_080 |
0.2444476758 |
- |
- |
PREDICTED: valine--tRNA ligase, mitochondrial isoform X2 [Jatropha curcas] |
| 7 |
Hb_000317_400 |
0.2448151615 |
transcription factor |
TF Family: zf-HD |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000096_130 |
0.2471588476 |
- |
- |
hypothetical protein 23 [Hevea brasiliensis] |
| 9 |
Hb_001865_080 |
0.2505068659 |
- |
- |
enoyl-CoA hydratase/isomerase family protein [Populus trichocarpa] |
| 10 |
Hb_021888_120 |
0.2512308418 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 11 |
Hb_002374_380 |
0.2513904172 |
- |
- |
Acyl-CoA N-acyltransferases (NAT) superfamily protein isoform 1 [Theobroma cacao] |
| 12 |
Hb_079268_010 |
0.2527772475 |
- |
- |
- |
| 13 |
Hb_004230_040 |
0.2533806511 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_115241_010 |
0.2545154539 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC [Jatropha curcas] |
| 15 |
Hb_002267_080 |
0.2546250192 |
- |
- |
PREDICTED: uncharacterized protein LOC105635094 [Jatropha curcas] |
| 16 |
Hb_000451_070 |
0.2555294767 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 17 |
Hb_000948_080 |
0.2558671636 |
- |
- |
PREDICTED: uncharacterized protein LOC105634692 [Jatropha curcas] |
| 18 |
Hb_000693_020 |
0.2589370005 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 19 |
Hb_002035_110 |
0.2595791472 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 20 |
Hb_002341_020 |
0.2598775458 |
- |
- |
PREDICTED: pyruvate decarboxylase 2 [Populus euphratica] |