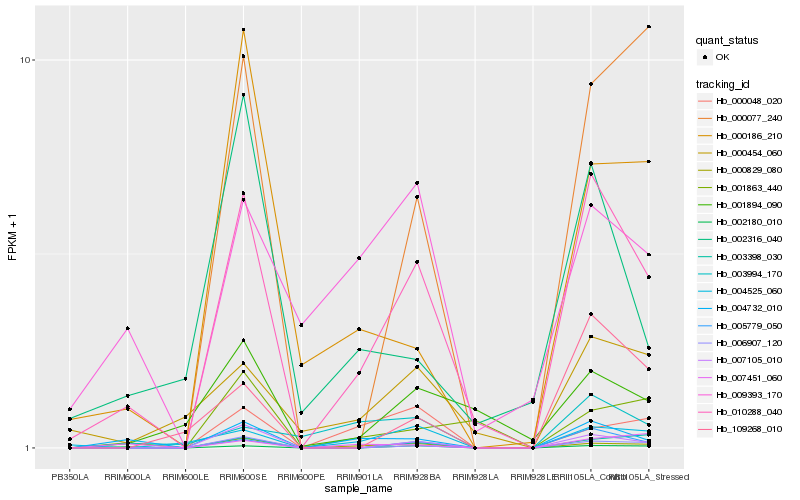
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010288_040 |
0.0 |
- |
- |
PREDICTED: transmembrane protein 205-like [Eucalyptus grandis] |
| 2 |
Hb_004732_010 |
0.212558947 |
- |
- |
- |
| 3 |
Hb_005779_050 |
0.2455146909 |
- |
- |
hypothetical protein JCGZ_14616 [Jatropha curcas] |
| 4 |
Hb_007105_010 |
0.2590979293 |
- |
- |
PREDICTED: uncharacterized protein LOC103455579 [Malus domestica] |
| 5 |
Hb_002180_010 |
0.262535797 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 6 |
Hb_000454_060 |
0.2642875025 |
- |
- |
Uncharacterized protein TCM_006778 [Theobroma cacao] |
| 7 |
Hb_006907_120 |
0.2679585524 |
- |
- |
PREDICTED: uncharacterized protein LOC103327233 [Prunus mume] |
| 8 |
Hb_004525_060 |
0.2721135465 |
- |
- |
- |
| 9 |
Hb_001894_090 |
0.282085573 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH90 [Jatropha curcas] |
| 10 |
Hb_000829_080 |
0.2856447815 |
- |
- |
PREDICTED: uncharacterized protein LOC105641027 [Jatropha curcas] |
| 11 |
Hb_003398_030 |
0.2907316135 |
- |
- |
PREDICTED: uncharacterized protein LOC104583892 [Brachypodium distachyon] |
| 12 |
Hb_000077_240 |
0.292459918 |
- |
- |
hypothetical protein PHAVU_011G114500g, partial [Phaseolus vulgaris] |
| 13 |
Hb_009393_170 |
0.294618523 |
- |
- |
PREDICTED: vestitone reductase-like [Jatropha curcas] |
| 14 |
Hb_000048_020 |
0.2952362738 |
- |
- |
- |
| 15 |
Hb_000186_210 |
0.2999414482 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 16 |
Hb_001863_440 |
0.3005207056 |
- |
- |
Nodulin-26, putative [Ricinus communis] |
| 17 |
Hb_002316_040 |
0.3041551934 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 18 |
Hb_007451_060 |
0.3064595172 |
- |
- |
BAHD family acyltransferase, clade V, putative [Theobroma cacao] |
| 19 |
Hb_003994_170 |
0.3091362701 |
- |
- |
Indole-3-acetic acid-induced protein ARG7, putative [Ricinus communis] |
| 20 |
Hb_109268_010 |
0.3134623706 |
- |
- |
PREDICTED: uncharacterized protein LOC105650977 [Jatropha curcas] |