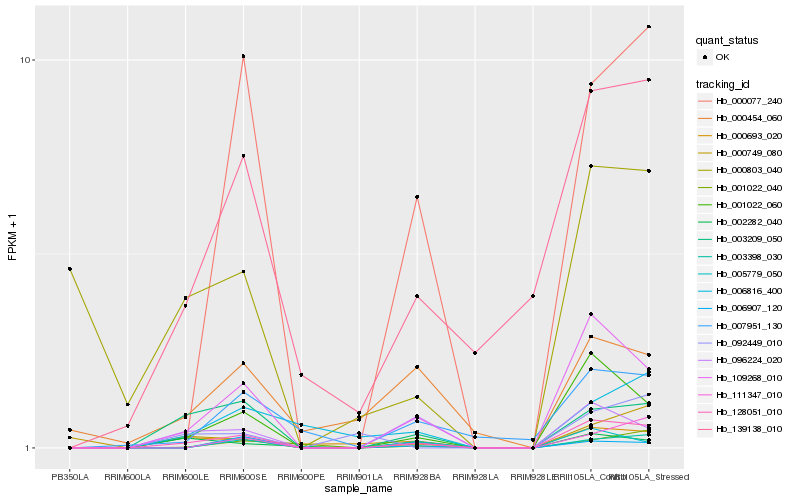
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_128051_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_109268_010 |
0.0975008884 |
- |
- |
PREDICTED: uncharacterized protein LOC105650977 [Jatropha curcas] |
| 3 |
Hb_001022_060 |
0.1309006304 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 4 |
Hb_003209_050 |
0.2232814255 |
- |
- |
hypothetical protein JCGZ_15683 [Jatropha curcas] |
| 5 |
Hb_000077_240 |
0.248703294 |
- |
- |
hypothetical protein PHAVU_011G114500g, partial [Phaseolus vulgaris] |
| 6 |
Hb_139138_010 |
0.2602250942 |
- |
- |
PREDICTED: uncharacterized protein LOC105650977 [Jatropha curcas] |
| 7 |
Hb_003398_030 |
0.2644604269 |
- |
- |
PREDICTED: uncharacterized protein LOC104583892 [Brachypodium distachyon] |
| 8 |
Hb_096224_020 |
0.2654773532 |
- |
- |
hypothetical protein JCGZ_06980 [Jatropha curcas] |
| 9 |
Hb_111347_010 |
0.2660860339 |
- |
- |
PREDICTED: uncharacterized protein LOC105781565 [Gossypium raimondii] |
| 10 |
Hb_005779_050 |
0.282487529 |
- |
- |
hypothetical protein JCGZ_14616 [Jatropha curcas] |
| 11 |
Hb_007951_130 |
0.282895543 |
desease resistance |
Gene Name: UBN2_3 |
JHL25H03.6 [Jatropha curcas] |
| 12 |
Hb_000693_020 |
0.289149113 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 13 |
Hb_002282_040 |
0.2934734183 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 14 |
Hb_000749_080 |
0.3058223521 |
- |
- |
Alpha-expansin 12 precursor, putative [Ricinus communis] |
| 15 |
Hb_006907_120 |
0.3060559203 |
- |
- |
PREDICTED: uncharacterized protein LOC103327233 [Prunus mume] |
| 16 |
Hb_001022_040 |
0.3074385169 |
- |
- |
PREDICTED: uncharacterized protein LOC105634753 [Jatropha curcas] |
| 17 |
Hb_006816_400 |
0.3085279996 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 18 |
Hb_000803_040 |
0.3140607741 |
- |
- |
- |
| 19 |
Hb_092449_010 |
0.3285418652 |
- |
- |
- |
| 20 |
Hb_000454_060 |
0.3289336086 |
- |
- |
Uncharacterized protein TCM_006778 [Theobroma cacao] |