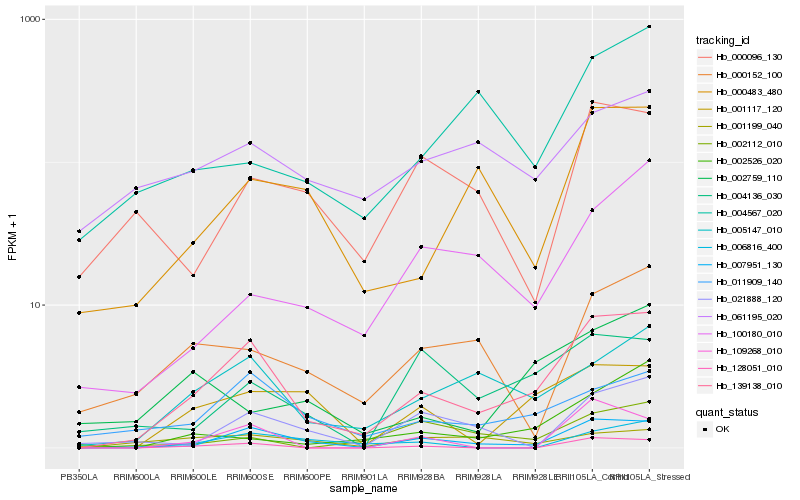
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_139138_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650977 [Jatropha curcas] |
| 2 |
Hb_007951_130 |
0.14647198 |
desease resistance |
Gene Name: UBN2_3 |
JHL25H03.6 [Jatropha curcas] |
| 3 |
Hb_005147_010 |
0.2040736702 |
- |
- |
PREDICTED: uncharacterized protein LOC105629670 [Jatropha curcas] |
| 4 |
Hb_011909_140 |
0.2224053669 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 5 |
Hb_001117_120 |
0.2427385953 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 6 |
Hb_000483_480 |
0.2515741064 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_128051_010 |
0.2602250942 |
- |
- |
- |
| 8 |
Hb_004136_030 |
0.2733582512 |
- |
- |
- |
| 9 |
Hb_000152_100 |
0.2750008418 |
- |
- |
PREDICTED: probable protein phosphatase 2C 75 isoform X2 [Jatropha curcas] |
| 10 |
Hb_021888_120 |
0.2751160984 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 11 |
Hb_100180_010 |
0.2787868781 |
- |
- |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 12 |
Hb_004567_020 |
0.2819680125 |
- |
- |
Small ubiquitin-related modifier 2 [Morus notabilis] |
| 13 |
Hb_002526_020 |
0.2825428121 |
- |
- |
- |
| 14 |
Hb_002112_010 |
0.2835529054 |
- |
- |
hypothetical protein POPTR_0001s45830g [Populus trichocarpa] |
| 15 |
Hb_061195_020 |
0.2836628225 |
- |
- |
PREDICTED: uncharacterized protein LOC101990594 [Microtus ochrogaster] |
| 16 |
Hb_006816_400 |
0.2843914261 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 17 |
Hb_000096_130 |
0.2874551294 |
- |
- |
hypothetical protein 23 [Hevea brasiliensis] |
| 18 |
Hb_109268_010 |
0.2884780857 |
- |
- |
PREDICTED: uncharacterized protein LOC105650977 [Jatropha curcas] |
| 19 |
Hb_001199_040 |
0.2892156467 |
- |
- |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_002759_110 |
0.2910960732 |
- |
- |
RNA-binding family protein, putative isoform 1 [Theobroma cacao] |