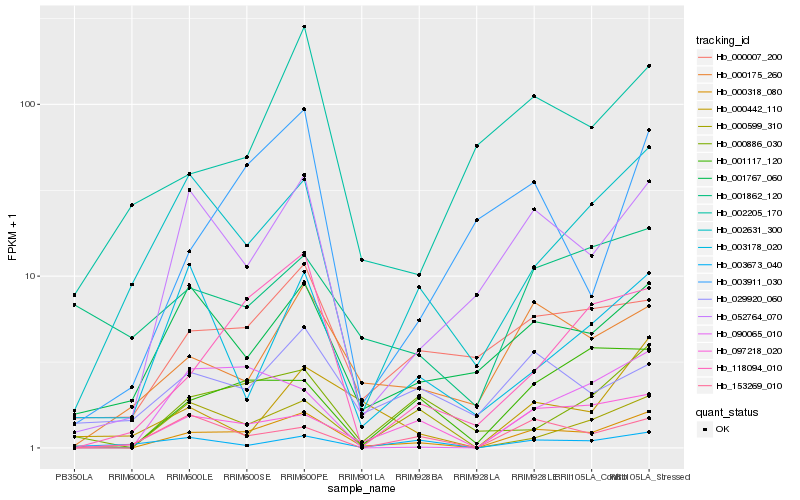
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000318_080 |
0.0 |
- |
- |
PREDICTED: la-related protein 6C [Jatropha curcas] |
| 2 |
Hb_052764_070 |
0.2172391009 |
- |
- |
PREDICTED: uncharacterized protein LOC105649473 [Jatropha curcas] |
| 3 |
Hb_097218_020 |
0.2244475393 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit E [Jatropha curcas] |
| 4 |
Hb_000007_200 |
0.2326846989 |
- |
- |
PREDICTED: choline/ethanolaminephosphotransferase 1 [Jatropha curcas] |
| 5 |
Hb_118094_010 |
0.2365461351 |
- |
- |
hypothetical protein POPTR_0011s06595g [Populus trichocarpa] |
| 6 |
Hb_001767_060 |
0.2377059129 |
- |
- |
PREDICTED: sulfate transporter 4.1, chloroplastic-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000886_030 |
0.2391302453 |
- |
- |
PREDICTED: uncharacterized protein LOC105113410 [Populus euphratica] |
| 8 |
Hb_000175_260 |
0.2399102691 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14-like [Populus euphratica] |
| 9 |
Hb_001117_120 |
0.2421237401 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 10 |
Hb_002631_300 |
0.24340033 |
- |
- |
PREDICTED: uncharacterized protein LOC105646181 [Jatropha curcas] |
| 11 |
Hb_003673_040 |
0.2614259051 |
- |
- |
dynamin family protein [Populus trichocarpa] |
| 12 |
Hb_029920_060 |
0.2635777653 |
- |
- |
PREDICTED: UPF0554 protein-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_001862_120 |
0.2680701327 |
- |
- |
PREDICTED: aspartic proteinase-like protein 2 [Jatropha curcas] |
| 14 |
Hb_002205_170 |
0.2683124671 |
- |
- |
PREDICTED: uncharacterized protein LOC105649179 [Jatropha curcas] |
| 15 |
Hb_153269_010 |
0.2690445228 |
- |
- |
hypothetical protein CICLE_v10029937mg [Citrus clementina] |
| 16 |
Hb_090065_010 |
0.2696420043 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 17 |
Hb_000599_310 |
0.2705012424 |
- |
- |
PREDICTED: pleckstrin homology domain-containing family A member 8-like [Jatropha curcas] |
| 18 |
Hb_000442_110 |
0.271542169 |
- |
- |
hypothetical protein POPTR_0010s18960g [Populus trichocarpa] |
| 19 |
Hb_003911_030 |
0.2736642274 |
- |
- |
glutamate dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_003178_020 |
0.2770453461 |
transcription factor |
TF Family: B3 |
conserved hypothetical protein [Ricinus communis] |