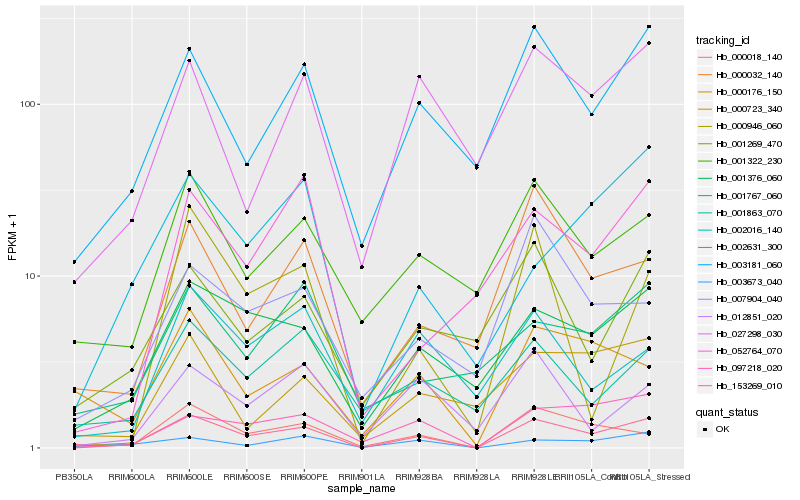
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_153269_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v10029937mg [Citrus clementina] |
| 2 |
Hb_001376_060 |
0.1873679307 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase abkC isoform X2 [Jatropha curcas] |
| 3 |
Hb_003181_060 |
0.1937555644 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Jatropha curcas] |
| 4 |
Hb_097218_020 |
0.1980627894 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit E [Jatropha curcas] |
| 5 |
Hb_000018_140 |
0.2044427689 |
- |
- |
PREDICTED: uncharacterized protein LOC105638239 [Jatropha curcas] |
| 6 |
Hb_003673_040 |
0.2156702772 |
- |
- |
dynamin family protein [Populus trichocarpa] |
| 7 |
Hb_027298_030 |
0.224005478 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 8 |
Hb_001322_230 |
0.2256920029 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 9 |
Hb_000032_140 |
0.2298652748 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001269_470 |
0.2325437361 |
- |
- |
PREDICTED: uncharacterized protein LOC105630313 [Jatropha curcas] |
| 11 |
Hb_052764_070 |
0.2331314987 |
- |
- |
PREDICTED: uncharacterized protein LOC105649473 [Jatropha curcas] |
| 12 |
Hb_001863_070 |
0.2374654372 |
- |
- |
Queuine tRNA-ribosyltransferase [Theobroma cacao] |
| 13 |
Hb_001767_060 |
0.238500415 |
- |
- |
PREDICTED: sulfate transporter 4.1, chloroplastic-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000723_340 |
0.2387048741 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG3 [Jatropha curcas] |
| 15 |
Hb_002631_300 |
0.2387913078 |
- |
- |
PREDICTED: uncharacterized protein LOC105646181 [Jatropha curcas] |
| 16 |
Hb_000946_060 |
0.2396458903 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_007904_040 |
0.2398845626 |
- |
- |
PREDICTED: uncharacterized protein LOC105644727 [Jatropha curcas] |
| 18 |
Hb_000176_150 |
0.2399339363 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_012851_020 |
0.240318549 |
- |
- |
PREDICTED: aminodeoxychorismate synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 20 |
Hb_002016_140 |
0.2405303037 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase BAH1-like 1 [Jatropha curcas] |