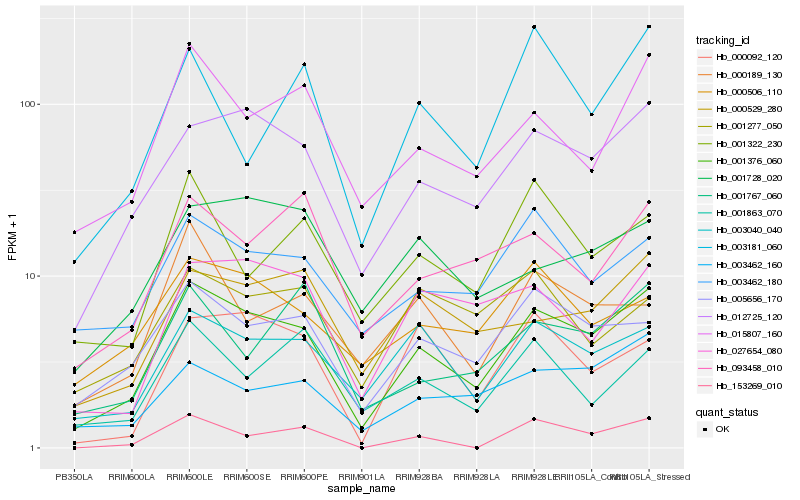
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001376_060 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase abkC isoform X2 [Jatropha curcas] |
| 2 |
Hb_012725_120 |
0.1086830433 |
- |
- |
PREDICTED: stress enhanced protein 2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_003040_040 |
0.1426124411 |
- |
- |
PREDICTED: protein SAWADEE HOMEODOMAIN HOMOLOG 1-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_005656_170 |
0.1430748829 |
- |
- |
PREDICTED: uncharacterized protein LOC105637447 isoform X1 [Jatropha curcas] |
| 5 |
Hb_015807_160 |
0.1474559154 |
- |
- |
PREDICTED: probable histone H2A.1 [Jatropha curcas] |
| 6 |
Hb_003462_160 |
0.1557541228 |
transcription factor |
TF Family: GNAT |
predicted protein [Arabidopsis lyrata subsp. lyrata] |
| 7 |
Hb_003462_180 |
0.1599313942 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable acetyltransferase NATA1-like [Jatropha curcas] |
| 8 |
Hb_000529_280 |
0.1612574678 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_027654_080 |
0.1665532182 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000506_110 |
0.167766586 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK11 [Jatropha curcas] |
| 11 |
Hb_093458_010 |
0.1711876408 |
- |
- |
hypothetical protein CISIN_1g0277592mg [Citrus sinensis] |
| 12 |
Hb_001322_230 |
0.1767060753 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 13 |
Hb_000092_120 |
0.1803262316 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 4 homolog A-like [Populus euphratica] |
| 14 |
Hb_001767_060 |
0.1803335719 |
- |
- |
PREDICTED: sulfate transporter 4.1, chloroplastic-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_001728_020 |
0.1818478775 |
- |
- |
PREDICTED: trans-1,2-dihydrobenzene-1,2-diol dehydrogenase [Jatropha curcas] |
| 16 |
Hb_001863_070 |
0.1866303406 |
- |
- |
Queuine tRNA-ribosyltransferase [Theobroma cacao] |
| 17 |
Hb_000189_130 |
0.1868732852 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATA, chloroplastic [Jatropha curcas] |
| 18 |
Hb_003181_060 |
0.1869501264 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Jatropha curcas] |
| 19 |
Hb_153269_010 |
0.1873679307 |
- |
- |
hypothetical protein CICLE_v10029937mg [Citrus clementina] |
| 20 |
Hb_001277_050 |
0.1875489437 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |