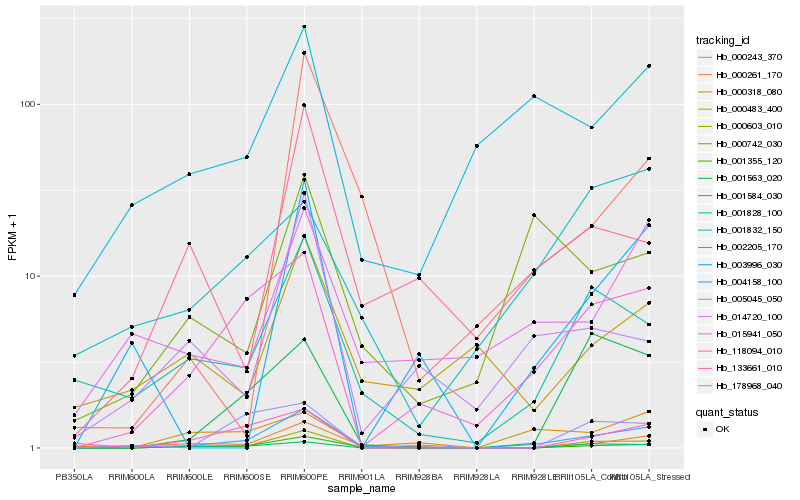
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004158_100 |
0.0 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2 [Jatropha curcas] |
| 2 |
Hb_001355_120 |
0.2088272289 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_000603_010 |
0.2100981297 |
- |
- |
PREDICTED: transmembrane protein 87A-like [Nelumbo nucifera] |
| 4 |
Hb_118094_010 |
0.2111611175 |
- |
- |
hypothetical protein POPTR_0011s06595g [Populus trichocarpa] |
| 5 |
Hb_000261_170 |
0.238162646 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 6 |
Hb_001828_100 |
0.2389125352 |
- |
- |
PREDICTED: death-associated protein kinase dapk-1 [Jatropha curcas] |
| 7 |
Hb_001584_030 |
0.2697000712 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like [Nelumbo nucifera] |
| 8 |
Hb_015941_050 |
0.2704442537 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001563_020 |
0.271931975 |
transcription factor |
TF Family: TRAF |
aberrant large forked product, putative [Ricinus communis] |
| 10 |
Hb_003996_030 |
0.2792197713 |
- |
- |
- |
| 11 |
Hb_002205_170 |
0.2861207276 |
- |
- |
PREDICTED: uncharacterized protein LOC105649179 [Jatropha curcas] |
| 12 |
Hb_000318_080 |
0.2882938607 |
- |
- |
PREDICTED: la-related protein 6C [Jatropha curcas] |
| 13 |
Hb_133661_010 |
0.2917291158 |
- |
- |
hypothetical protein JCGZ_11734 [Jatropha curcas] |
| 14 |
Hb_000243_370 |
0.2962978362 |
- |
- |
hypothetical protein VITISV_010022 [Vitis vinifera] |
| 15 |
Hb_178968_040 |
0.2972197254 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005045_050 |
0.3003087585 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 3 [Jatropha curcas] |
| 17 |
Hb_014720_100 |
0.3021888448 |
- |
- |
PREDICTED: probable boron transporter 2 [Jatropha curcas] |
| 18 |
Hb_000483_400 |
0.3042318175 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000742_030 |
0.3069207784 |
- |
- |
PREDICTED: uncharacterized protein LOC105636851 [Jatropha curcas] |
| 20 |
Hb_001832_150 |
0.3075311181 |
- |
- |
PREDICTED: uncharacterized protein LOC105637882 [Jatropha curcas] |