| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
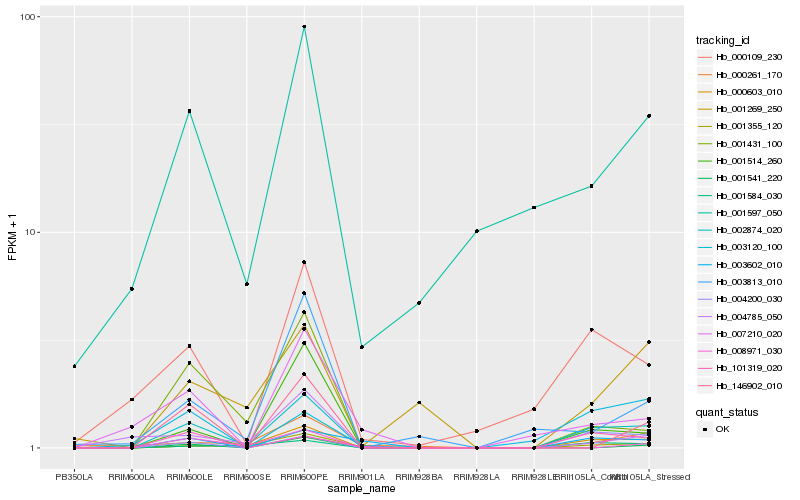
Hb_002874_020 |
0.0 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105637459 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000603_010 |
0.2198333899 |
- |
- |
PREDICTED: transmembrane protein 87A-like [Nelumbo nucifera] |
| 3 |
Hb_001584_030 |
0.2456356715 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like [Nelumbo nucifera] |
| 4 |
Hb_001355_120 |
0.2618299876 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_000109_230 |
0.2619306243 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001514_260 |
0.2664535867 |
- |
- |
- |
| 7 |
Hb_001431_100 |
0.2676728739 |
- |
- |
PREDICTED: probable protein phosphatase 2C 27 [Jatropha curcas] |
| 8 |
Hb_004200_030 |
0.2794336559 |
- |
- |
PREDICTED: SWR1 complex subunit 6-like isoform X1 [Citrus sinensis] |
| 9 |
Hb_003602_010 |
0.2838620464 |
- |
- |
serine hydroxymethyltransferase, partial [Populus alba] |
| 10 |
Hb_101319_020 |
0.2841320814 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 11 |
Hb_001541_220 |
0.2985951502 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_0793640 [Ricinus communis] |
| 12 |
Hb_001597_050 |
0.3000761851 |
- |
- |
- |
| 13 |
Hb_146902_010 |
0.3013350654 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001269_250 |
0.3028918414 |
- |
- |
PREDICTED: factor of DNA methylation 1-like [Jatropha curcas] |
| 15 |
Hb_003813_010 |
0.3035858896 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BEE 1-like [Jatropha curcas] |
| 16 |
Hb_008971_030 |
0.3080170185 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003120_100 |
0.3082684417 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 18 |
Hb_007210_020 |
0.3152485728 |
- |
- |
PREDICTED: hexaprenyldihydroxybenzoate methyltransferase, mitochondrial-like [Solanum tuberosum] |
| 19 |
Hb_000261_170 |
0.3190833518 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 20 |
Hb_004785_050 |
0.3201743645 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |