| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
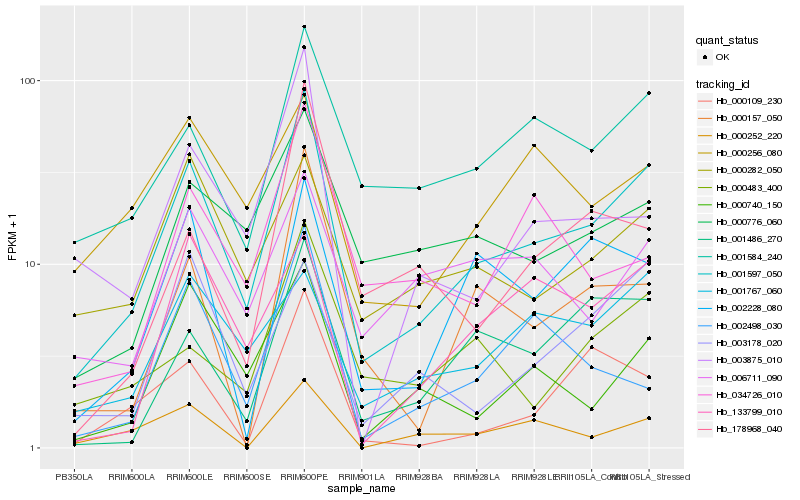
Hb_001597_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_001584_240 |
0.1825232481 |
- |
- |
ADP-ribosylation factor [Gossypium barbadense] |
| 3 |
Hb_000740_150 |
0.192456029 |
- |
- |
PREDICTED: uncharacterized protein LOC105629998 [Jatropha curcas] |
| 4 |
Hb_001486_270 |
0.1948138605 |
- |
- |
Ann10 [Manihot esculenta] |
| 5 |
Hb_000776_060 |
0.1960734978 |
- |
- |
histone H4 [Zea mays] |
| 6 |
Hb_000483_400 |
0.1998072312 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1, mitochondrial [Jatropha curcas] |
| 7 |
Hb_003875_010 |
0.2030108224 |
- |
- |
hypothetical protein POPTR_0002s14490g, partial [Populus trichocarpa] |
| 8 |
Hb_000157_050 |
0.2043688945 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002228_080 |
0.2058276059 |
- |
- |
hypothetical protein JCGZ_12924 [Jatropha curcas] |
| 10 |
Hb_000282_050 |
0.2084620269 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 11 |
Hb_003178_020 |
0.211892719 |
transcription factor |
TF Family: B3 |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_133799_010 |
0.2142906275 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006711_090 |
0.2145692692 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |
| 14 |
Hb_000109_230 |
0.2147672327 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000252_220 |
0.2159930627 |
- |
- |
PREDICTED: copper amine oxidase 1-like isoform X2 [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_001767_060 |
0.2173999077 |
- |
- |
PREDICTED: sulfate transporter 4.1, chloroplastic-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_178968_040 |
0.2192470626 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_034726_010 |
0.2195134152 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X2 [Jatropha curcas] |
| 19 |
Hb_002498_030 |
0.2228913026 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000256_080 |
0.2237676823 |
- |
- |
PREDICTED: deSI-like protein At4g17486 [Jatropha curcas] |