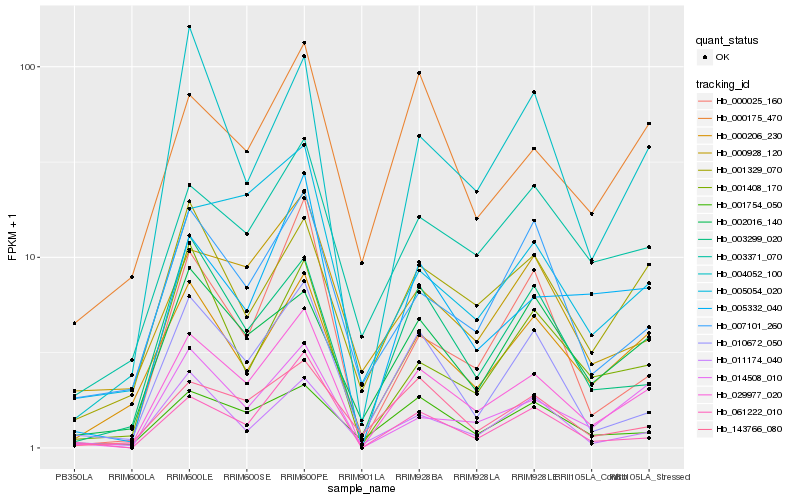
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029977_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640153 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005332_040 |
0.1413851329 |
- |
- |
hypothetical protein RCOM_0530890 [Ricinus communis] |
| 3 |
Hb_000206_230 |
0.1423167876 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_014508_010 |
0.1480334658 |
- |
- |
PREDICTED: uncharacterized protein LOC105629998 [Jatropha curcas] |
| 5 |
Hb_010672_050 |
0.1503287123 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X3 [Jatropha curcas] |
| 6 |
Hb_004052_100 |
0.152947533 |
- |
- |
PREDICTED: protein SPIRAL1-like 3 [Jatropha curcas] |
| 7 |
Hb_003299_020 |
0.1621897049 |
- |
- |
PREDICTED: uncharacterized protein LOC105649783 [Jatropha curcas] |
| 8 |
Hb_061222_010 |
0.1634237167 |
- |
- |
hypothetical protein RCOM_0012650 [Ricinus communis] |
| 9 |
Hb_001329_070 |
0.1659530344 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 10 |
Hb_000025_160 |
0.1662061177 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003371_070 |
0.1669769027 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 12 |
Hb_143766_080 |
0.1693352156 |
- |
- |
PREDICTED: uncharacterized protein LOC105637797 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007101_260 |
0.1703979234 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 14 |
Hb_000175_470 |
0.172746652 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 15 |
Hb_001754_050 |
0.1731303167 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_001408_170 |
0.1734398667 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 2 [Jatropha curcas] |
| 17 |
Hb_002016_140 |
0.1737434332 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase BAH1-like 1 [Jatropha curcas] |
| 18 |
Hb_005054_020 |
0.1747730437 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 19 |
Hb_011174_040 |
0.1760256475 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000928_120 |
0.1764778906 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |