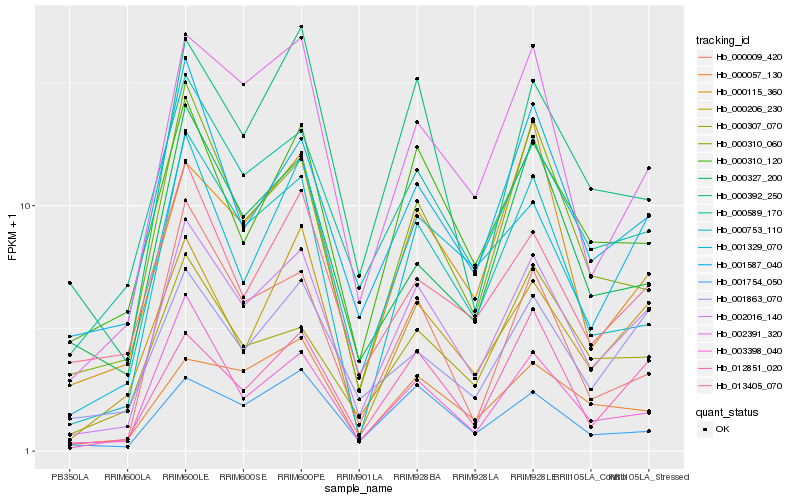
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002016_140 |
0.0 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase BAH1-like 1 [Jatropha curcas] |
| 2 |
Hb_002391_320 |
0.109337519 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 3 |
Hb_001329_070 |
0.1113830051 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 4 |
Hb_000589_170 |
0.1184321099 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 5 |
Hb_012851_020 |
0.1215436087 |
- |
- |
PREDICTED: aminodeoxychorismate synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 6 |
Hb_000753_110 |
0.1224244136 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM1 isoform X6 [Jatropha curcas] |
| 7 |
Hb_000206_230 |
0.1224359417 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000310_120 |
0.1232392633 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |
| 9 |
Hb_001587_040 |
0.1248416554 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001863_070 |
0.126264821 |
- |
- |
Queuine tRNA-ribosyltransferase [Theobroma cacao] |
| 11 |
Hb_003398_040 |
0.1263427489 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001754_050 |
0.1295879133 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_000310_060 |
0.1352896197 |
- |
- |
hypothetical protein JCGZ_20793 [Jatropha curcas] |
| 14 |
Hb_000057_130 |
0.1357942634 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 15 |
Hb_000307_070 |
0.1366777889 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 16 |
Hb_000392_250 |
0.1372500088 |
- |
- |
PREDICTED: HVA22-like protein a [Jatropha curcas] |
| 17 |
Hb_000115_360 |
0.1411611146 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 18 |
Hb_013405_070 |
0.1422686167 |
- |
- |
ferric-chelate reductase, putative [Ricinus communis] |
| 19 |
Hb_000009_420 |
0.1436307097 |
- |
- |
PREDICTED: uncharacterized protein LOC105639614 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000327_200 |
0.1455908288 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |