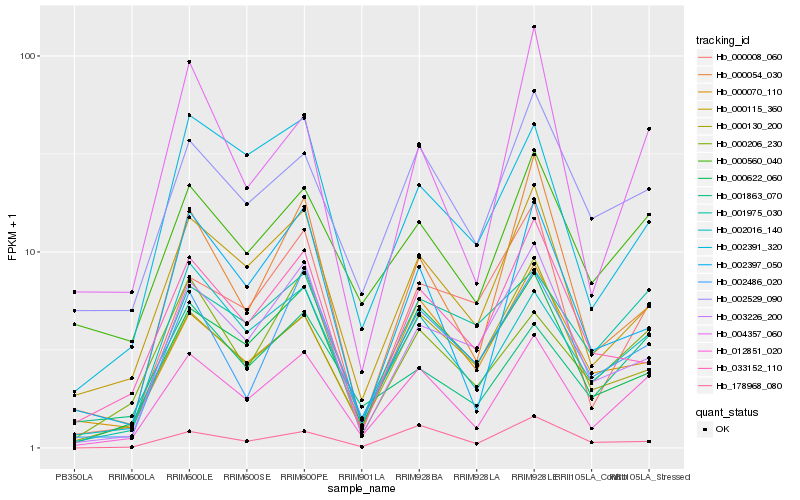
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012851_020 |
0.0 |
- |
- |
PREDICTED: aminodeoxychorismate synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 2 |
Hb_002016_140 |
0.1215436087 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase BAH1-like 1 [Jatropha curcas] |
| 3 |
Hb_000115_360 |
0.1274681434 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 4 |
Hb_000622_060 |
0.1371145652 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000560_040 |
0.1373901057 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002529_090 |
0.1414945436 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 7 |
Hb_002486_020 |
0.1443138573 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_000130_200 |
0.1480583095 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |
| 9 |
Hb_001975_030 |
0.1552135443 |
- |
- |
4-diphosphocytidyl-2-c-methyl-d-erythritol kinase, partial [Plectranthus barbatus] |
| 10 |
Hb_000070_110 |
0.1562183202 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002397_050 |
0.1568930137 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4 isoform X2 [Jatropha curcas] |
| 12 |
Hb_178968_080 |
0.1586291999 |
- |
- |
PREDICTED: serine/threonine-protein kinase OSR1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_003226_200 |
0.1591509975 |
- |
- |
magnesium/proton exchanger, putative [Ricinus communis] |
| 14 |
Hb_002391_320 |
0.1593950862 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 15 |
Hb_000206_230 |
0.1600210469 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001863_070 |
0.160180138 |
- |
- |
Queuine tRNA-ribosyltransferase [Theobroma cacao] |
| 17 |
Hb_004357_060 |
0.1603697428 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 18 |
Hb_000054_030 |
0.1617423785 |
- |
- |
hypothetical protein POPTR_0010s17680g [Populus trichocarpa] |
| 19 |
Hb_033152_110 |
0.1648344461 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN3 [Jatropha curcas] |
| 20 |
Hb_000008_060 |
0.1687677764 |
- |
- |
protein binding protein, putative [Ricinus communis] |