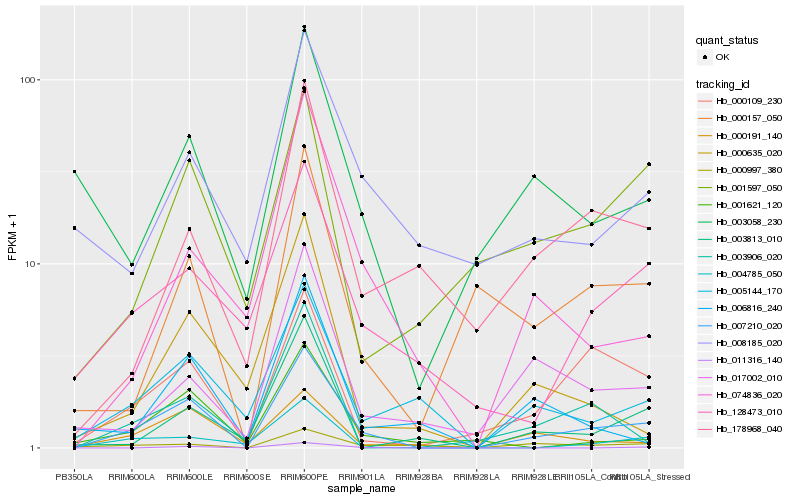
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007210_020 |
0.0 |
- |
- |
PREDICTED: hexaprenyldihydroxybenzoate methyltransferase, mitochondrial-like [Solanum tuberosum] |
| 2 |
Hb_000109_230 |
0.2313559824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000191_140 |
0.2436445205 |
- |
- |
PREDICTED: growth-regulating factor 1-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_005144_170 |
0.251427171 |
- |
- |
PREDICTED: shugoshin-1-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_004785_050 |
0.2533685818 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 6 |
Hb_128473_010 |
0.2624214161 |
- |
- |
- |
| 7 |
Hb_000157_050 |
0.262653314 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001621_120 |
0.26498218 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 9 |
Hb_000997_380 |
0.2658405484 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |
| 10 |
Hb_003906_020 |
0.2681022 |
- |
- |
PREDICTED: uncharacterized protein LOC105648535 [Jatropha curcas] |
| 11 |
Hb_017002_010 |
0.2697875751 |
- |
- |
PREDICTED: expansin-A15-like [Jatropha curcas] |
| 12 |
Hb_074836_020 |
0.2732591269 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_178968_040 |
0.2736382169 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003813_010 |
0.2748475233 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BEE 1-like [Jatropha curcas] |
| 15 |
Hb_000635_020 |
0.2791614193 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_011316_140 |
0.2795489568 |
- |
- |
PREDICTED: uncharacterized protein LOC105129591 [Populus euphratica] |
| 17 |
Hb_001597_050 |
0.2821740173 |
- |
- |
- |
| 18 |
Hb_003058_230 |
0.2832305091 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 19 |
Hb_008185_020 |
0.2848315451 |
- |
- |
RING-H2 finger protein ATL3J, putative [Ricinus communis] |
| 20 |
Hb_006816_240 |
0.2850074527 |
- |
- |
PREDICTED: uncharacterized protein LOC105647443 [Jatropha curcas] |