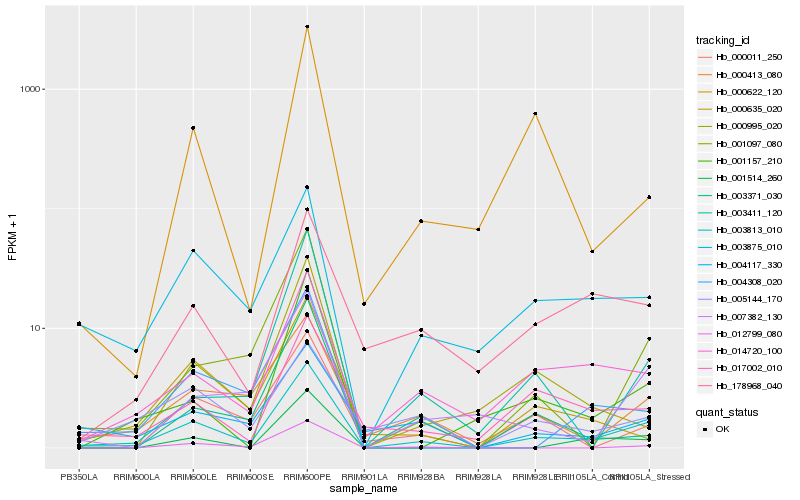
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003813_010 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BEE 1-like [Jatropha curcas] |
| 2 |
Hb_014720_100 |
0.19071267 |
- |
- |
PREDICTED: probable boron transporter 2 [Jatropha curcas] |
| 3 |
Hb_001097_080 |
0.2042777402 |
transcription factor |
TF Family: MYB-related |
triptychon and cpc, putative [Ricinus communis] |
| 4 |
Hb_000413_080 |
0.2069586127 |
- |
- |
- |
| 5 |
Hb_003875_010 |
0.2110103465 |
- |
- |
hypothetical protein POPTR_0002s14490g, partial [Populus trichocarpa] |
| 6 |
Hb_017002_010 |
0.2118934144 |
- |
- |
PREDICTED: expansin-A15-like [Jatropha curcas] |
| 7 |
Hb_007382_130 |
0.2225044078 |
- |
- |
glycerophosphodiester phosphodiesterase, putative [Ricinus communis] |
| 8 |
Hb_000011_250 |
0.223147874 |
- |
- |
PREDICTED: RING-H2 finger protein ATL3-like [Jatropha curcas] |
| 9 |
Hb_012799_080 |
0.2243802184 |
transcription factor |
TF Family: MYB |
Myb domain protein 18, putative [Theobroma cacao] |
| 10 |
Hb_004117_330 |
0.228473808 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001157_210 |
0.2284894225 |
- |
- |
hypothetical protein CISIN_1g038260mg [Citrus sinensis] |
| 12 |
Hb_003411_120 |
0.2353829523 |
- |
- |
hypothetical protein POPTR_0010s13670g [Populus trichocarpa] |
| 13 |
Hb_003371_030 |
0.2361217366 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004308_020 |
0.2361934292 |
- |
- |
hypothetical protein POPTR_0005s15590g [Populus trichocarpa] |
| 15 |
Hb_178968_040 |
0.2366658643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000635_020 |
0.236957101 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001514_260 |
0.2412643971 |
- |
- |
- |
| 18 |
Hb_000622_120 |
0.2412776515 |
- |
- |
hypothetical protein [Hevea brasiliensis] |
| 19 |
Hb_005144_170 |
0.2432042167 |
- |
- |
PREDICTED: shugoshin-1-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_000995_020 |
0.243219259 |
- |
- |
conserved hypothetical protein [Ricinus communis] |