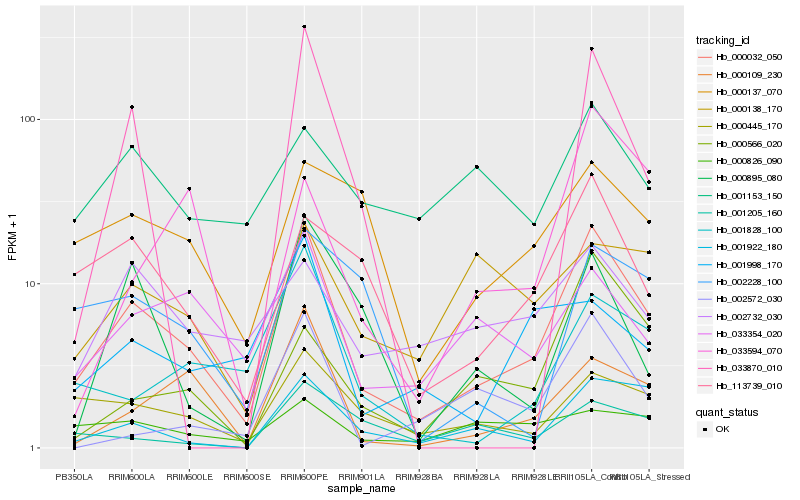
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000032_050 |
0.0 |
- |
- |
hypothetical protein RCOM_1691130 [Ricinus communis] |
| 2 |
Hb_001922_180 |
0.2272704016 |
- |
- |
hevamine-A precursor, putative [Ricinus communis] |
| 3 |
Hb_000445_170 |
0.2292124994 |
- |
- |
PREDICTED: probable methyltransferase PMT10 [Jatropha curcas] |
| 4 |
Hb_113739_010 |
0.2319632833 |
- |
- |
hypothetical protein JCGZ_06379 [Jatropha curcas] |
| 5 |
Hb_000109_230 |
0.2357694645 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_033354_020 |
0.241460145 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_001828_100 |
0.2446579987 |
- |
- |
PREDICTED: death-associated protein kinase dapk-1 [Jatropha curcas] |
| 8 |
Hb_000137_070 |
0.2633606658 |
- |
- |
PREDICTED: uncharacterized protein LOC105644263 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002732_030 |
0.2642956265 |
transcription factor |
TF Family: AP2 |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002572_030 |
0.266330091 |
- |
- |
PREDICTED: diacylglycerol kinase theta [Jatropha curcas] |
| 11 |
Hb_001998_170 |
0.2670187661 |
- |
- |
BnaC07g08230D [Brassica napus] |
| 12 |
Hb_000895_080 |
0.2687119221 |
- |
- |
PREDICTED: acyl-CoA--sterol O-acyltransferase 1-like [Jatropha curcas] |
| 13 |
Hb_000566_020 |
0.2713607653 |
- |
- |
hypothetical protein POPTR_0001s13520g [Populus trichocarpa] |
| 14 |
Hb_033870_010 |
0.275275622 |
- |
- |
PREDICTED: uncharacterized protein LOC104101109 isoform X1 [Nicotiana tomentosiformis] |
| 15 |
Hb_002228_100 |
0.2761036116 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 16 |
Hb_001153_150 |
0.281381934 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
HMG-CoA synthase 2 [Hevea brasiliensis] |
| 17 |
Hb_000138_170 |
0.2815020127 |
- |
- |
putative plastid-lipid-associated protein, partial [Hevea brasiliensis] |
| 18 |
Hb_033594_070 |
0.2821105078 |
- |
- |
PREDICTED: uncharacterized protein LOC105787685 isoform X1 [Gossypium raimondii] |
| 19 |
Hb_001205_160 |
0.282775082 |
- |
- |
hypothetical protein POPTR_0005s12560g [Populus trichocarpa] |
| 20 |
Hb_000826_090 |
0.2832460819 |
- |
- |
PREDICTED: cationic peroxidase 1-like [Populus euphratica] |