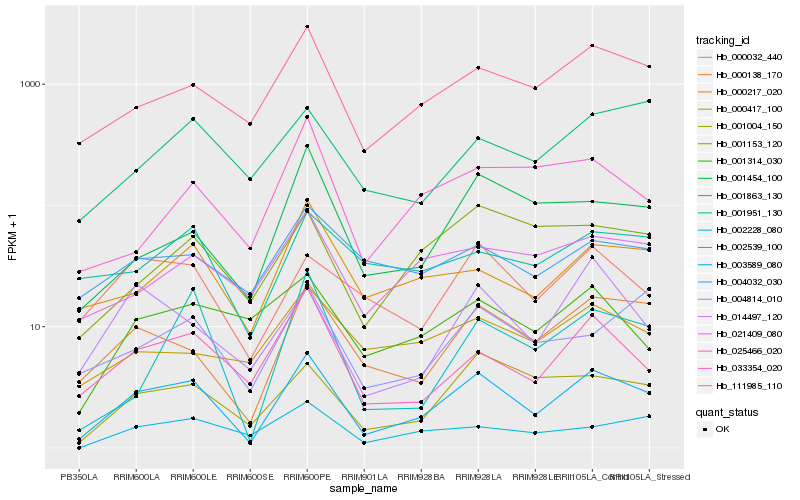
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003589_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105107863 [Populus euphratica] |
| 2 |
Hb_000138_170 |
0.1661130828 |
- |
- |
putative plastid-lipid-associated protein, partial [Hevea brasiliensis] |
| 3 |
Hb_033354_020 |
0.1768577233 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_111985_110 |
0.1770895406 |
- |
- |
thioredoxin H-type 3 [Hevea brasiliensis] |
| 5 |
Hb_001004_150 |
0.1824968879 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-3-like [Jatropha curcas] |
| 6 |
Hb_000217_020 |
0.1951509546 |
- |
- |
AMME syndrome candidateprotein 1 protein, putative [Ricinus communis] |
| 7 |
Hb_000032_440 |
0.1959984616 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_001454_100 |
0.2021503227 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 9 |
Hb_002228_080 |
0.2036107837 |
- |
- |
hypothetical protein JCGZ_12924 [Jatropha curcas] |
| 10 |
Hb_014497_120 |
0.2084418857 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
RecName: Full=3-hydroxy-3-methylglutaryl-coenzyme A reductase 1; Short=HMG-CoA reductase 1 [Hevea brasiliensis] |
| 11 |
Hb_004814_010 |
0.2098199897 |
- |
- |
PREDICTED: uncharacterized protein LOC105633486 [Jatropha curcas] |
| 12 |
Hb_002539_100 |
0.2110020888 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 iota [Jatropha curcas] |
| 13 |
Hb_021409_080 |
0.2116691376 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |
| 14 |
Hb_001153_120 |
0.2140155354 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 15 |
Hb_001314_030 |
0.2173374432 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 16 |
Hb_001951_130 |
0.2205868956 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 17 |
Hb_004032_030 |
0.2222113049 |
- |
- |
PREDICTED: RNA-binding protein 24-like isoform X1 [Populus euphratica] |
| 18 |
Hb_025466_020 |
0.2224658638 |
- |
- |
tubulin beta-6 [Brassica oleracea var. italica] |
| 19 |
Hb_000417_100 |
0.223697596 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 12-like [Jatropha curcas] |
| 20 |
Hb_001863_130 |
0.2237299676 |
- |
- |
hypothetical protein ZEAMMB73_433257, partial [Zea mays] |