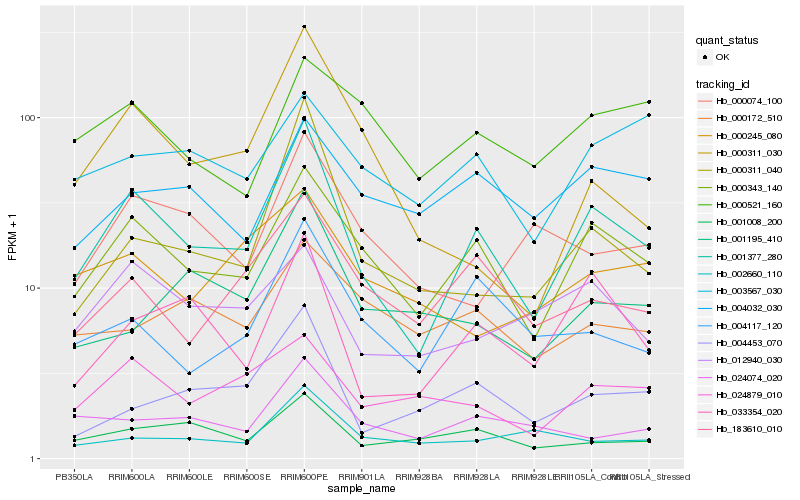
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001377_280 |
0.0 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Populus euphratica] |
| 2 |
Hb_000343_140 |
0.1247314519 |
- |
- |
PREDICTED: uncharacterized protein LOC105634966 [Jatropha curcas] |
| 3 |
Hb_033354_020 |
0.1700048834 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_000311_040 |
0.1832960786 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |
| 5 |
Hb_183610_010 |
0.1925811674 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 6 |
Hb_004453_070 |
0.1937200231 |
transcription factor |
TF Family: CPP |
PREDICTED: CRC domain-containing protein TSO1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_004117_120 |
0.1966822922 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 8 |
Hb_024879_010 |
0.1995747491 |
- |
- |
unknown [Glycine max] |
| 9 |
Hb_001008_200 |
0.1997328496 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC103961552 [Pyrus x bretschneideri] |
| 10 |
Hb_012940_030 |
0.2032706273 |
- |
- |
PREDICTED: uncharacterized protein LOC105639708 [Jatropha curcas] |
| 11 |
Hb_024074_020 |
0.213997523 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g09600 [Jatropha curcas] |
| 12 |
Hb_001195_410 |
0.2157944808 |
- |
- |
PREDICTED: deoxyuridine 5'-triphosphate nucleotidohydrolase [Jatropha curcas] |
| 13 |
Hb_000074_100 |
0.218134289 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000311_030 |
0.2185097966 |
- |
- |
hypothetical protein B456_002G227600 [Gossypium raimondii] |
| 15 |
Hb_004032_030 |
0.2203040611 |
- |
- |
PREDICTED: RNA-binding protein 24-like isoform X1 [Populus euphratica] |
| 16 |
Hb_002660_110 |
0.2238878216 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003567_030 |
0.2258014959 |
- |
- |
PREDICTED: nifU-like protein 4, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000245_080 |
0.2261867786 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-15 [Jatropha curcas] |
| 19 |
Hb_000521_160 |
0.2281777979 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000172_510 |
0.2300339129 |
- |
- |
hypothetical protein B456_007G243100 [Gossypium raimondii] |