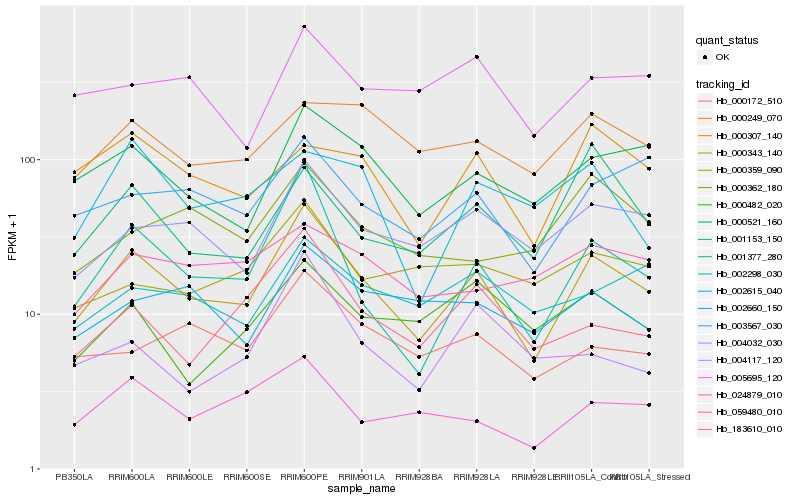
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000343_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634966 [Jatropha curcas] |
| 2 |
Hb_001377_280 |
0.1247314519 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Populus euphratica] |
| 3 |
Hb_000521_160 |
0.1419484298 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004032_030 |
0.1433192358 |
- |
- |
PREDICTED: RNA-binding protein 24-like isoform X1 [Populus euphratica] |
| 5 |
Hb_003567_030 |
0.1564500254 |
- |
- |
PREDICTED: nifU-like protein 4, mitochondrial [Jatropha curcas] |
| 6 |
Hb_024879_010 |
0.1568375867 |
- |
- |
unknown [Glycine max] |
| 7 |
Hb_183610_010 |
0.1578365472 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 8 |
Hb_000172_510 |
0.1628784902 |
- |
- |
hypothetical protein B456_007G243100 [Gossypium raimondii] |
| 9 |
Hb_002660_150 |
0.1705905973 |
- |
- |
PREDICTED: uncharacterized protein LOC105642207 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005695_120 |
0.1718356221 |
- |
- |
Pollen-specific protein C13 precursor, putative [Ricinus communis] |
| 11 |
Hb_001153_150 |
0.1723424258 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
HMG-CoA synthase 2 [Hevea brasiliensis] |
| 12 |
Hb_000307_140 |
0.173759328 |
- |
- |
PREDICTED: 24-methylenesterol C-methyltransferase 2 [Jatropha curcas] |
| 13 |
Hb_000249_070 |
0.1739538573 |
- |
- |
PREDICTED: probable calcium-binding protein CML35 [Jatropha curcas] |
| 14 |
Hb_002298_030 |
0.1749146171 |
- |
- |
PREDICTED: coenzyme Q-binding protein COQ10 homolog, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000482_020 |
0.1759363413 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 16 |
Hb_000359_090 |
0.1772890567 |
- |
- |
PREDICTED: persulfide dioxygenase ETHE1 homolog, mitochondrial-like [Malus domestica] |
| 17 |
Hb_000362_180 |
0.1792005578 |
- |
- |
Pdap1 [Gossypium arboreum] |
| 18 |
Hb_004117_120 |
0.1816896784 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 19 |
Hb_059480_010 |
0.1828356293 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 20 |
Hb_002615_040 |
0.1829306708 |
- |
- |
PREDICTED: uncharacterized protein LOC105110770 [Populus euphratica] |