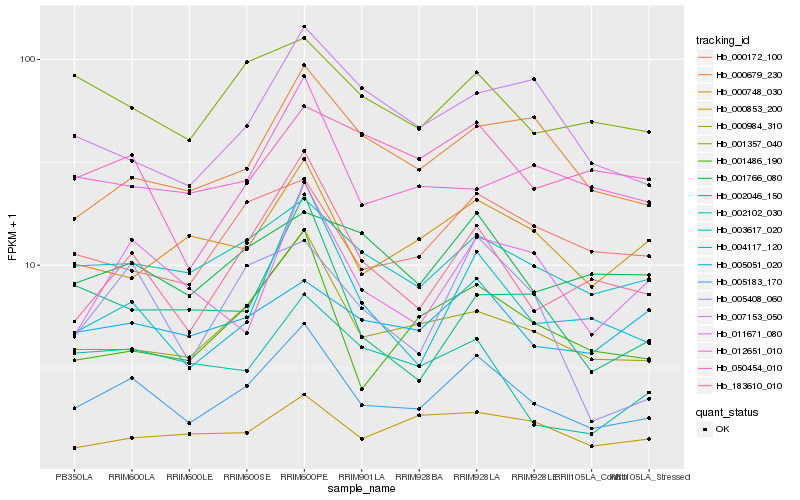
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005183_170 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g04860 [Jatropha curcas] |
| 2 |
Hb_183610_010 |
0.1137266251 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 3 |
Hb_000984_310 |
0.119698101 |
- |
- |
Hydrolase, hydrolyzing O-glycosyl compounds, putative [Theobroma cacao] |
| 4 |
Hb_003617_020 |
0.1320487132 |
- |
- |
PREDICTED: uncharacterized protein LOC105638179 [Jatropha curcas] |
| 5 |
Hb_004117_120 |
0.1325123535 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 6 |
Hb_001486_190 |
0.1325182313 |
- |
- |
hypothetical protein JCGZ_02702 [Jatropha curcas] |
| 7 |
Hb_011671_080 |
0.1494330695 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase TDR [Jatropha curcas] |
| 8 |
Hb_000172_100 |
0.1501102418 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 2, peroxisomal [Jatropha curcas] |
| 9 |
Hb_000679_230 |
0.1510689712 |
- |
- |
PREDICTED: serine/threonine-protein kinase PRP4 homolog [Jatropha curcas] |
| 10 |
Hb_005408_060 |
0.1514429717 |
- |
- |
PREDICTED: uncharacterized protein LOC105648045 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001766_080 |
0.1518282203 |
- |
- |
PREDICTED: uncharacterized protein LOC105644125 [Jatropha curcas] |
| 12 |
Hb_001357_040 |
0.1523826806 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 13 |
Hb_000748_030 |
0.1526657647 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_005051_020 |
0.1560234548 |
- |
- |
hypothetical protein JCGZ_20345 [Jatropha curcas] |
| 15 |
Hb_000853_200 |
0.1593580179 |
- |
- |
PREDICTED: uncharacterized protein LOC105642574 [Jatropha curcas] |
| 16 |
Hb_012651_010 |
0.1616683341 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 17 |
Hb_002102_030 |
0.1627999639 |
- |
- |
- |
| 18 |
Hb_007153_050 |
0.16375953 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 8 [Jatropha curcas] |
| 19 |
Hb_002046_150 |
0.1647304361 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g15080 [Jatropha curcas] |
| 20 |
Hb_050454_010 |
0.1655418327 |
- |
- |
PREDICTED: uncharacterized protein LOC105630497 [Jatropha curcas] |