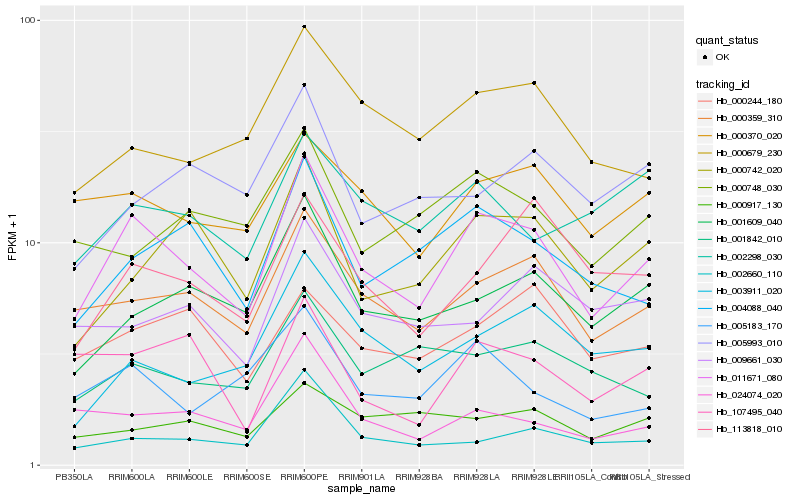
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011671_080 |
0.0 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase TDR [Jatropha curcas] |
| 2 |
Hb_000359_310 |
0.1182370273 |
- |
- |
hypothetical protein JCGZ_03934 [Jatropha curcas] |
| 3 |
Hb_004088_040 |
0.1425823949 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' iota isoform [Jatropha curcas] |
| 4 |
Hb_001609_040 |
0.1444479406 |
- |
- |
hypothetical protein POPTR_0017s05650g [Populus trichocarpa] |
| 5 |
Hb_000370_020 |
0.1454232539 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000742_020 |
0.1470994756 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 7 |
Hb_005183_170 |
0.1494330695 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g04860 [Jatropha curcas] |
| 8 |
Hb_000679_230 |
0.151527383 |
- |
- |
PREDICTED: serine/threonine-protein kinase PRP4 homolog [Jatropha curcas] |
| 9 |
Hb_113818_010 |
0.1556580488 |
- |
- |
Xyloglucan galactosyltransferase KATAMARI1, putative [Ricinus communis] |
| 10 |
Hb_003911_020 |
0.1591678808 |
- |
- |
PREDICTED: uncharacterized protein LOC105121332 [Populus euphratica] |
| 11 |
Hb_001842_010 |
0.1596884642 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 12 |
Hb_002298_030 |
0.1607822427 |
- |
- |
PREDICTED: coenzyme Q-binding protein COQ10 homolog, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000244_180 |
0.1630600961 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_024074_020 |
0.1637368262 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g09600 [Jatropha curcas] |
| 15 |
Hb_000748_030 |
0.1644005871 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_107495_040 |
0.165874394 |
- |
- |
PREDICTED: 12-oxophytodienoate reductase 2-like [Citrus sinensis] |
| 17 |
Hb_000917_130 |
0.166269697 |
- |
- |
PREDICTED: uncharacterized protein LOC105635730 [Jatropha curcas] |
| 18 |
Hb_002660_110 |
0.1665053489 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_009661_030 |
0.1676763461 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like [Jatropha curcas] |
| 20 |
Hb_005993_010 |
0.1680759559 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |