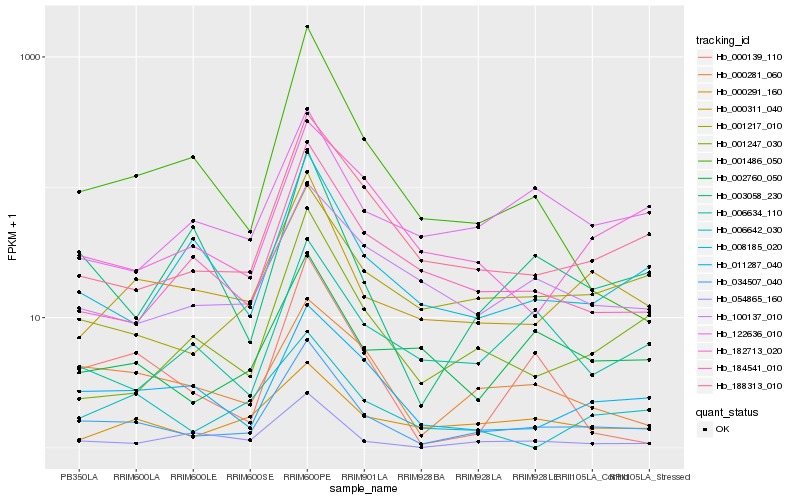
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_034507_040 |
0.0 |
- |
- |
peptide transporter, putative [Ricinus communis] |
| 2 |
Hb_188313_010 |
0.1725907774 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 3 |
Hb_000311_040 |
0.1744076606 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |
| 4 |
Hb_054865_160 |
0.185872492 |
- |
- |
PREDICTED: D-arabinono-1,4-lactone oxidase-like [Jatropha curcas] |
| 5 |
Hb_001217_010 |
0.1920200478 |
- |
- |
hypothetical protein JCGZ_04022 [Jatropha curcas] |
| 6 |
Hb_011287_040 |
0.1931017709 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_001247_030 |
0.1989387953 |
- |
- |
- |
| 8 |
Hb_000281_060 |
0.200002479 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_008185_020 |
0.2034514865 |
- |
- |
RING-H2 finger protein ATL3J, putative [Ricinus communis] |
| 10 |
Hb_002760_050 |
0.2049972509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_184541_010 |
0.205418276 |
- |
- |
PREDICTED: selT-like protein [Jatropha curcas] |
| 12 |
Hb_001486_050 |
0.2106642878 |
- |
- |
PREDICTED: arabinogalactan peptide 13-like [Jatropha curcas] |
| 13 |
Hb_003058_230 |
0.2163952027 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 14 |
Hb_100137_010 |
0.2173138265 |
- |
- |
hypothetical protein JCGZ_17848 [Jatropha curcas] |
| 15 |
Hb_122636_010 |
0.2210810131 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 16 |
Hb_000139_110 |
0.2218659733 |
- |
- |
PREDICTED: mavicyanin [Jatropha curcas] |
| 17 |
Hb_006634_110 |
0.2223086158 |
- |
- |
BnaC04g40780D [Brassica napus] |
| 18 |
Hb_182713_020 |
0.2257474559 |
- |
- |
PREDICTED: vesicle-associated membrane protein 727 [Setaria italica] |
| 19 |
Hb_000291_160 |
0.226746702 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |
| 20 |
Hb_006642_030 |
0.2290196434 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Jatropha curcas] |