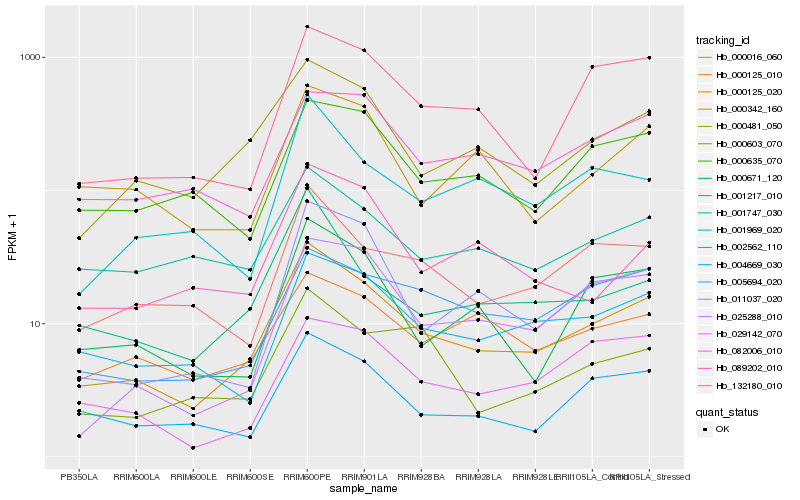
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000125_020 |
0.0 |
transcription factor |
TF Family: mTERF |
- |
| 2 |
Hb_000481_050 |
0.1184623408 |
- |
- |
PREDICTED: uncharacterized protein LOC105636546 [Jatropha curcas] |
| 3 |
Hb_002562_110 |
0.1374994041 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 4 |
Hb_004669_030 |
0.1410508681 |
- |
- |
- |
| 5 |
Hb_132180_010 |
0.1411765975 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 6 |
Hb_000671_120 |
0.1413541308 |
- |
- |
hypothetical protein JCGZ_23777 [Jatropha curcas] |
| 7 |
Hb_000016_060 |
0.1425422666 |
- |
- |
mads box protein, putative [Ricinus communis] |
| 8 |
Hb_011037_020 |
0.1469710718 |
- |
- |
unknown [Picea sitchensis] |
| 9 |
Hb_001217_010 |
0.1481494539 |
- |
- |
hypothetical protein JCGZ_04022 [Jatropha curcas] |
| 10 |
Hb_005694_020 |
0.149842188 |
- |
- |
PREDICTED: ORM1-like protein 2 [Gossypium raimondii] |
| 11 |
Hb_025288_010 |
0.1531575513 |
- |
- |
PREDICTED: protein unc-50 homolog [Jatropha curcas] |
| 12 |
Hb_001969_020 |
0.1564949912 |
- |
- |
hypothetical protein POPTR_0010s04700g [Populus trichocarpa] |
| 13 |
Hb_000635_070 |
0.157291563 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 14 |
Hb_001747_030 |
0.1582448463 |
- |
- |
hypothetical protein F383_08210 [Gossypium arboreum] |
| 15 |
Hb_000342_160 |
0.1584085488 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 16 |
Hb_082006_010 |
0.1587851099 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000603_070 |
0.1604709038 |
- |
- |
- |
| 18 |
Hb_000125_010 |
0.1611344558 |
transcription factor |
TF Family: mTERF |
- |
| 19 |
Hb_029142_070 |
0.1613086461 |
- |
- |
- |
| 20 |
Hb_089202_010 |
0.1618681253 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Nicotiana sylvestris] |