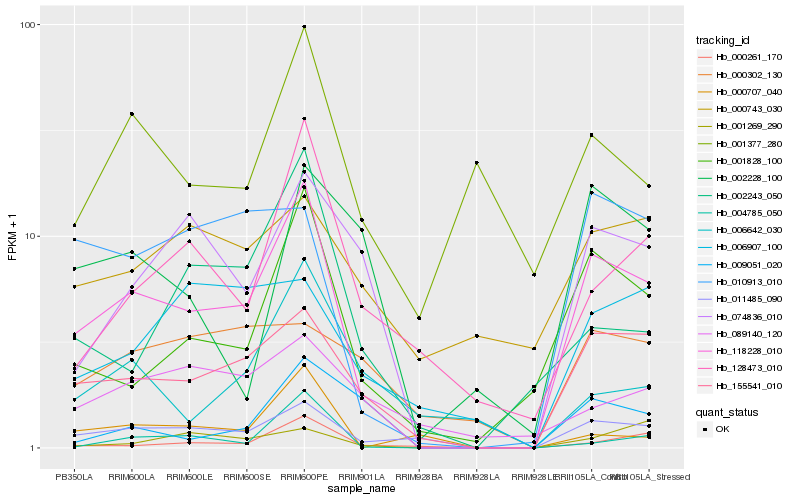
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_118228_010 |
0.0 |
- |
- |
PREDICTED: pirin-like protein [Jatropha curcas] |
| 2 |
Hb_155541_010 |
0.1772272316 |
- |
- |
- |
| 3 |
Hb_011485_090 |
0.1857234647 |
- |
- |
- |
| 4 |
Hb_001828_100 |
0.2023346905 |
- |
- |
PREDICTED: death-associated protein kinase dapk-1 [Jatropha curcas] |
| 5 |
Hb_074836_010 |
0.2410139537 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_010913_010 |
0.2430093625 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 7 |
Hb_000261_170 |
0.2498168703 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 8 |
Hb_128473_010 |
0.2535067834 |
- |
- |
- |
| 9 |
Hb_004785_050 |
0.2717355566 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 10 |
Hb_000707_040 |
0.2731499453 |
- |
- |
Uncharacterized protein TCM_021829 [Theobroma cacao] |
| 11 |
Hb_002243_050 |
0.2731649209 |
- |
- |
PREDICTED: ion channel DMI1-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_001377_280 |
0.2755439315 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Populus euphratica] |
| 13 |
Hb_009051_020 |
0.2814318142 |
- |
- |
Thioredoxin O1 [Morus notabilis] |
| 14 |
Hb_002228_100 |
0.2858451914 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 15 |
Hb_006907_100 |
0.2865915922 |
- |
- |
PREDICTED: tropinone reductase-like 3 [Jatropha curcas] |
| 16 |
Hb_001269_290 |
0.2950636249 |
- |
- |
PREDICTED: uncharacterized protein LOC103438859 [Malus domestica] |
| 17 |
Hb_006642_030 |
0.2952086094 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Jatropha curcas] |
| 18 |
Hb_000743_030 |
0.2956551958 |
- |
- |
NmrA-like negative transcriptional regulator family protein [Theobroma cacao] |
| 19 |
Hb_089140_120 |
0.2972721655 |
- |
- |
PREDICTED: MOB kinase activator-like 1 [Jatropha curcas] |
| 20 |
Hb_000302_130 |
0.3014522507 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 1 [Jatropha curcas] |