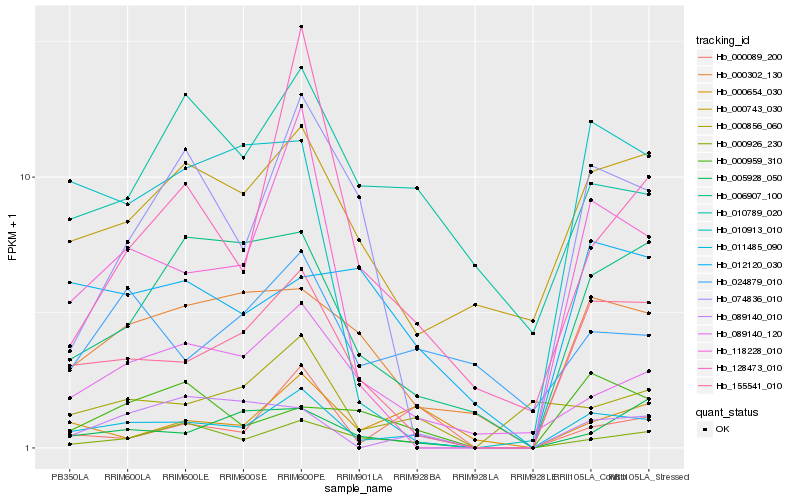
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011485_090 |
0.0 |
- |
- |
- |
| 2 |
Hb_155541_010 |
0.1615499467 |
- |
- |
- |
| 3 |
Hb_118228_010 |
0.1857234647 |
- |
- |
PREDICTED: pirin-like protein [Jatropha curcas] |
| 4 |
Hb_000926_230 |
0.2127728566 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_006907_100 |
0.2169077492 |
- |
- |
PREDICTED: tropinone reductase-like 3 [Jatropha curcas] |
| 6 |
Hb_000302_130 |
0.218410853 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 1 [Jatropha curcas] |
| 7 |
Hb_089140_120 |
0.2205924662 |
- |
- |
PREDICTED: MOB kinase activator-like 1 [Jatropha curcas] |
| 8 |
Hb_000743_030 |
0.2252551452 |
- |
- |
NmrA-like negative transcriptional regulator family protein [Theobroma cacao] |
| 9 |
Hb_000856_060 |
0.241419676 |
- |
- |
- |
| 10 |
Hb_010913_010 |
0.2417314093 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 11 |
Hb_000654_030 |
0.2430449694 |
- |
- |
- |
| 12 |
Hb_000089_200 |
0.246665802 |
- |
- |
hypothetical protein VITISV_019292 [Vitis vinifera] |
| 13 |
Hb_010789_020 |
0.2468054004 |
- |
- |
ubiquitin-conjugating enzyme e2S, putative [Ricinus communis] |
| 14 |
Hb_128473_010 |
0.2485732732 |
- |
- |
- |
| 15 |
Hb_074836_010 |
0.2497817116 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_024879_010 |
0.250090344 |
- |
- |
unknown [Glycine max] |
| 17 |
Hb_089140_010 |
0.2504590506 |
- |
- |
- |
| 18 |
Hb_005928_050 |
0.2513395387 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 19 |
Hb_000959_310 |
0.2540893957 |
- |
- |
PREDICTED: WD repeat domain-containing protein 83 [Jatropha curcas] |
| 20 |
Hb_012120_030 |
0.2547462089 |
- |
- |
- |