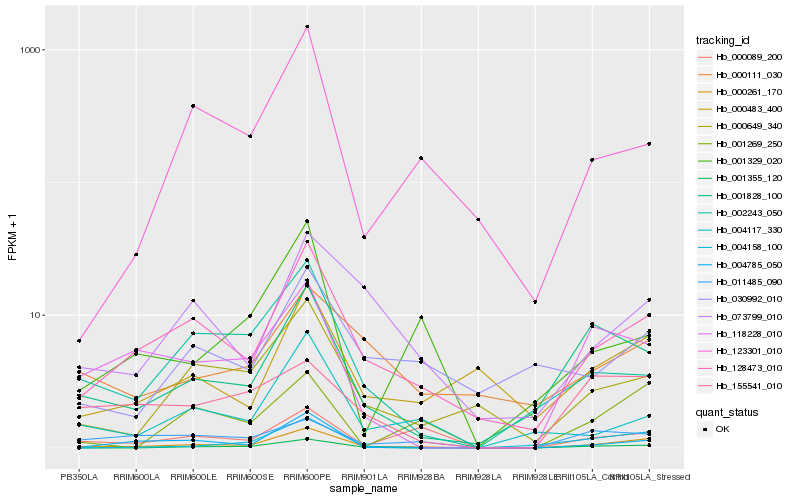
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_170 |
0.0 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 2 |
Hb_128473_010 |
0.1612420496 |
- |
- |
- |
| 3 |
Hb_001355_120 |
0.2184783654 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_000649_340 |
0.2339904715 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic [Jatropha curcas] |
| 5 |
Hb_001828_100 |
0.2355030744 |
- |
- |
PREDICTED: death-associated protein kinase dapk-1 [Jatropha curcas] |
| 6 |
Hb_004158_100 |
0.238162646 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2 [Jatropha curcas] |
| 7 |
Hb_004785_050 |
0.2411740055 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 8 |
Hb_000483_400 |
0.2419770195 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1, mitochondrial [Jatropha curcas] |
| 9 |
Hb_073799_010 |
0.2432780464 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001269_250 |
0.2472514288 |
- |
- |
PREDICTED: factor of DNA methylation 1-like [Jatropha curcas] |
| 11 |
Hb_118228_010 |
0.2498168703 |
- |
- |
PREDICTED: pirin-like protein [Jatropha curcas] |
| 12 |
Hb_004117_330 |
0.2499473085 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000089_200 |
0.2561972846 |
- |
- |
hypothetical protein VITISV_019292 [Vitis vinifera] |
| 14 |
Hb_002243_050 |
0.2598578112 |
- |
- |
PREDICTED: ion channel DMI1-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_123301_010 |
0.2655342886 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_011485_090 |
0.2683667208 |
- |
- |
- |
| 17 |
Hb_000111_030 |
0.2704703299 |
- |
- |
PREDICTED: uncharacterized protein LOC104094030 [Nicotiana tomentosiformis] |
| 18 |
Hb_030992_010 |
0.2728264607 |
- |
- |
acetylornithine aminotransferase, putative [Ricinus communis] |
| 19 |
Hb_155541_010 |
0.2746285616 |
- |
- |
- |
| 20 |
Hb_001329_020 |
0.2757649252 |
- |
- |
PREDICTED: probable boron transporter 2 [Populus euphratica] |