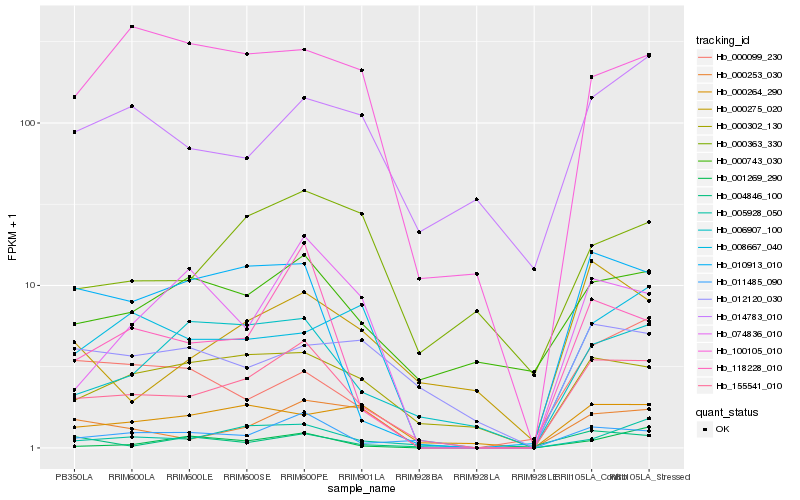
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_155541_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_011485_090 |
0.1615499467 |
- |
- |
- |
| 3 |
Hb_118228_010 |
0.1772272316 |
- |
- |
PREDICTED: pirin-like protein [Jatropha curcas] |
| 4 |
Hb_005928_050 |
0.1898569954 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 5 |
Hb_000302_130 |
0.1904302063 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 1 [Jatropha curcas] |
| 6 |
Hb_010913_010 |
0.1954391829 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 7 |
Hb_006907_100 |
0.1960468384 |
- |
- |
PREDICTED: tropinone reductase-like 3 [Jatropha curcas] |
| 8 |
Hb_000253_030 |
0.2050204036 |
- |
- |
hypothetical protein JCGZ_23039 [Jatropha curcas] |
| 9 |
Hb_000743_030 |
0.211383404 |
- |
- |
NmrA-like negative transcriptional regulator family protein [Theobroma cacao] |
| 10 |
Hb_074836_010 |
0.217663737 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000264_290 |
0.2207027303 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit RPB2 [Jatropha curcas] |
| 12 |
Hb_000275_020 |
0.2301909896 |
- |
- |
PREDICTED: uncharacterized protein LOC105632656 [Jatropha curcas] |
| 13 |
Hb_004846_100 |
0.2346313684 |
- |
- |
Na(+)/H(+) antiporter, putative [Ricinus communis] |
| 14 |
Hb_000363_330 |
0.2385787863 |
- |
- |
- |
| 15 |
Hb_100105_010 |
0.239104574 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 16 |
Hb_012120_030 |
0.2416836301 |
- |
- |
- |
| 17 |
Hb_014783_010 |
0.2425558721 |
- |
- |
cold-inducible RNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_001269_290 |
0.2459108247 |
- |
- |
PREDICTED: uncharacterized protein LOC103438859 [Malus domestica] |
| 19 |
Hb_008667_040 |
0.2466296831 |
- |
- |
glycerol-3-phosphate transporter family protein [Populus trichocarpa] |
| 20 |
Hb_000099_230 |
0.247706501 |
- |
- |
PREDICTED: F-box protein SKIP19-like [Gossypium raimondii] |