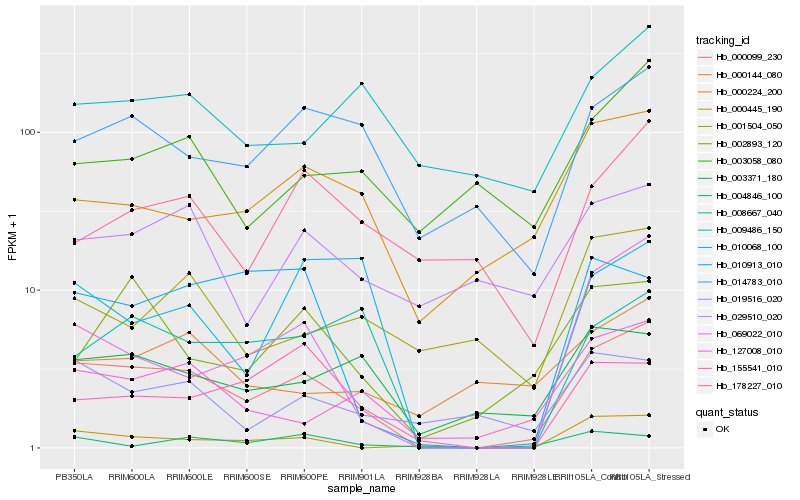
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000099_230 |
0.0 |
- |
- |
PREDICTED: F-box protein SKIP19-like [Gossypium raimondii] |
| 2 |
Hb_127008_010 |
0.1998003229 |
- |
- |
PREDICTED: uncharacterized protein LOC105629501 [Jatropha curcas] |
| 3 |
Hb_069022_010 |
0.2176299345 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_014783_010 |
0.2192722848 |
- |
- |
cold-inducible RNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_000445_190 |
0.2193772617 |
- |
- |
- |
| 6 |
Hb_003371_180 |
0.2255405665 |
- |
- |
- |
| 7 |
Hb_008667_040 |
0.2259439924 |
- |
- |
glycerol-3-phosphate transporter family protein [Populus trichocarpa] |
| 8 |
Hb_000224_200 |
0.2289376096 |
- |
- |
PREDICTED: uncharacterized protein LOC105640874 [Jatropha curcas] |
| 9 |
Hb_002893_120 |
0.2297853829 |
- |
- |
PREDICTED: uncharacterized protein LOC105637692 [Jatropha curcas] |
| 10 |
Hb_010913_010 |
0.2383952432 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 11 |
Hb_019516_020 |
0.2414597964 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_010068_100 |
0.2435369519 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |
| 13 |
Hb_155541_010 |
0.247706501 |
- |
- |
- |
| 14 |
Hb_029510_020 |
0.2482311301 |
- |
- |
hypothetical protein JCGZ_06306 [Jatropha curcas] |
| 15 |
Hb_004846_100 |
0.2517152742 |
- |
- |
Na(+)/H(+) antiporter, putative [Ricinus communis] |
| 16 |
Hb_009486_150 |
0.2543384871 |
- |
- |
60S ribosomal protein L37a-2 [Medicago truncatula] |
| 17 |
Hb_003058_080 |
0.2555135981 |
- |
- |
Chaperonin 10 isoform 1 [Theobroma cacao] |
| 18 |
Hb_178227_010 |
0.2563527537 |
- |
- |
Mitochondrial import inner membrane translocase subunit Tim13, putative [Ricinus communis] |
| 19 |
Hb_000144_080 |
0.2582699466 |
- |
- |
PREDICTED: uncharacterized protein LOC104609774 [Nelumbo nucifera] |
| 20 |
Hb_001504_050 |
0.2589232209 |
- |
- |
hypothetical protein POPTR_0001s14840g [Populus trichocarpa] |