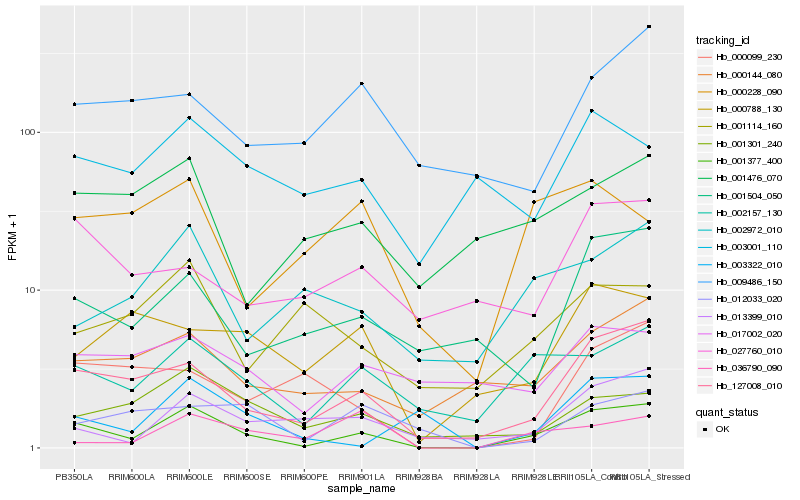
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001377_400 |
0.0 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 2 |
Hb_127008_010 |
0.1822280299 |
- |
- |
PREDICTED: uncharacterized protein LOC105629501 [Jatropha curcas] |
| 3 |
Hb_002157_130 |
0.222658215 |
- |
- |
- |
| 4 |
Hb_000144_080 |
0.2550431643 |
- |
- |
PREDICTED: uncharacterized protein LOC104609774 [Nelumbo nucifera] |
| 5 |
Hb_013399_010 |
0.2594245018 |
- |
- |
PREDICTED: uncharacterized protein LOC105631404 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001504_050 |
0.2677711584 |
- |
- |
hypothetical protein POPTR_0001s14840g [Populus trichocarpa] |
| 7 |
Hb_001301_240 |
0.2699094779 |
- |
- |
- |
| 8 |
Hb_002972_010 |
0.2754421765 |
- |
- |
PREDICTED: magnesium-dependent phosphatase 1 [Jatropha curcas] |
| 9 |
Hb_003322_010 |
0.2777366628 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_017002_020 |
0.2863949284 |
- |
- |
PREDICTED: uncharacterized protein LOC105650411 [Jatropha curcas] |
| 11 |
Hb_001476_070 |
0.2901204587 |
- |
- |
MinE protein [Manihot esculenta] |
| 12 |
Hb_000788_130 |
0.2905689112 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Jatropha curcas] |
| 13 |
Hb_027760_010 |
0.2914364701 |
- |
- |
PREDICTED: uncharacterized protein LOC105630460 [Jatropha curcas] |
| 14 |
Hb_036790_090 |
0.2927903104 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_012033_020 |
0.2928268023 |
- |
- |
- |
| 16 |
Hb_009486_150 |
0.2952666928 |
- |
- |
60S ribosomal protein L37a-2 [Medicago truncatula] |
| 17 |
Hb_003001_110 |
0.2960860524 |
- |
- |
oleoyl-acyl carrier protein thioesterase 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000099_230 |
0.2965743184 |
- |
- |
PREDICTED: F-box protein SKIP19-like [Gossypium raimondii] |
| 19 |
Hb_001114_160 |
0.2972705152 |
- |
- |
PREDICTED: beta carbonic anhydrase 5, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000228_090 |
0.2977884409 |
- |
- |
PREDICTED: protein PAM68, chloroplastic [Jatropha curcas] |