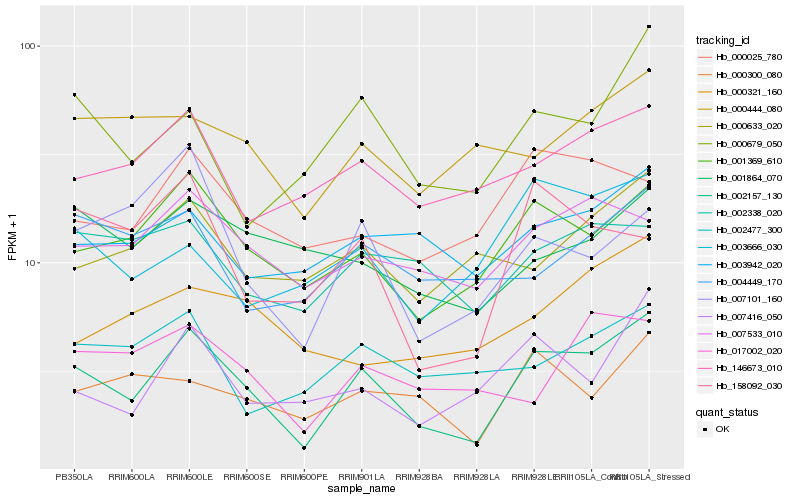
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002157_130 |
0.0 |
- |
- |
- |
| 2 |
Hb_001369_610 |
0.1409538423 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000679_050 |
0.1672805138 |
- |
- |
Alba DNA/RNA-binding protein [Theobroma cacao] |
| 4 |
Hb_007416_050 |
0.1693253051 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 5 [Jatropha curcas] |
| 5 |
Hb_146673_010 |
0.1750284687 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 6 |
Hb_007533_010 |
0.1751937188 |
transcription factor |
TF Family: bZIP |
G-box-binding factor, putative [Ricinus communis] |
| 7 |
Hb_000300_080 |
0.176968829 |
- |
- |
PREDICTED: probable S-sulfocysteine synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_003942_020 |
0.1787742786 |
- |
- |
PREDICTED: multiple RNA-binding domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000025_780 |
0.1809309387 |
- |
- |
PREDICTED: sanguinarine reductase [Jatropha curcas] |
| 10 |
Hb_003666_030 |
0.1827294514 |
- |
- |
Thiol:disulfide interchange protein txlA, putative [Ricinus communis] |
| 11 |
Hb_002338_020 |
0.1832221237 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR34A [Jatropha curcas] |
| 12 |
Hb_000633_020 |
0.1842466273 |
- |
- |
PREDICTED: uncharacterized protein LOC105630002 [Jatropha curcas] |
| 13 |
Hb_002477_300 |
0.1843368899 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_017002_020 |
0.1851580134 |
- |
- |
PREDICTED: uncharacterized protein LOC105650411 [Jatropha curcas] |
| 15 |
Hb_000444_080 |
0.1857053182 |
- |
- |
hypothetical protein POPTR_0019s10500g [Populus trichocarpa] |
| 16 |
Hb_007101_160 |
0.1865910915 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004449_170 |
0.1868130357 |
- |
- |
PREDICTED: uncharacterized protein LOC105631285 isoform X1 [Jatropha curcas] |
| 18 |
Hb_158092_030 |
0.1872954385 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP19, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001864_070 |
0.1878056149 |
- |
- |
PREDICTED: probable histone H2A.3 [Jatropha curcas] |
| 20 |
Hb_000321_160 |
0.1903578645 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 9 isoform X1 [Jatropha curcas] |