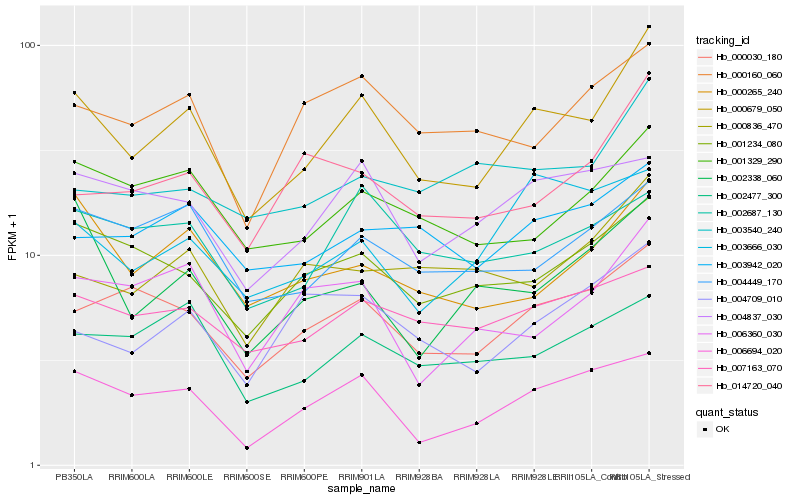
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000679_050 |
0.0 |
- |
- |
Alba DNA/RNA-binding protein [Theobroma cacao] |
| 2 |
Hb_000030_180 |
0.1220950085 |
- |
- |
PREDICTED: uncharacterized protein LOC105632645 isoform X1 [Jatropha curcas] |
| 3 |
Hb_006694_020 |
0.1253042054 |
- |
- |
PREDICTED: auxin response factor 2-like isoform X2 [Nicotiana tomentosiformis] |
| 4 |
Hb_001329_290 |
0.13234089 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 5 |
Hb_000265_240 |
0.1324406254 |
- |
- |
PREDICTED: uncharacterized protein LOC105643590 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001234_080 |
0.133636578 |
- |
- |
PREDICTED: eyes absent homolog 2-like isoform X2 [Musa acuminata subsp. malaccensis] |
| 7 |
Hb_004449_170 |
0.1367083271 |
- |
- |
PREDICTED: uncharacterized protein LOC105631285 isoform X1 [Jatropha curcas] |
| 8 |
Hb_007163_070 |
0.1397855821 |
- |
- |
PREDICTED: tRNA-dihydrouridine(20) synthase [NAD(P)+]-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_003942_020 |
0.1412736165 |
- |
- |
PREDICTED: multiple RNA-binding domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004837_030 |
0.142062546 |
- |
- |
PREDICTED: mitochondrial tRNA-specific 2-thiouridylase 1 [Jatropha curcas] |
| 11 |
Hb_004709_010 |
0.1424885268 |
- |
- |
hypothetical protein B456_007G097300 [Gossypium raimondii] |
| 12 |
Hb_002477_300 |
0.1432214063 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_000160_060 |
0.1439300426 |
- |
- |
unknown [Populus trichocarpa] |
| 14 |
Hb_014720_040 |
0.1443631061 |
- |
- |
PREDICTED: 50S ribosomal protein L12, chloroplastic [Jatropha curcas] |
| 15 |
Hb_006360_030 |
0.14511821 |
- |
- |
- |
| 16 |
Hb_002687_130 |
0.1472579185 |
- |
- |
PREDICTED: uncharacterized protein LOC105631153 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003540_240 |
0.1476176497 |
- |
- |
PREDICTED: ribosomal RNA-processing protein 17 [Jatropha curcas] |
| 18 |
Hb_000836_470 |
0.1492954035 |
- |
- |
- |
| 19 |
Hb_003666_030 |
0.1500455979 |
- |
- |
Thiol:disulfide interchange protein txlA, putative [Ricinus communis] |
| 20 |
Hb_002338_060 |
0.1503323487 |
- |
- |
2Fe-2S ferredoxin [Glycine soja] |